Simulation-Study
Simulation-Study.RmdSimulate Data
Covariates
t = seq(0, 1, length.out = 100)
n = 500
components = 3
mean_funs = list(
function(t) -2*(t-0.5)^2 + 5,
function(t) 3*(t-0.75)^3 - 5
)
eigen_funs_list = list(
list(
function(t) sin(2*pi*t),
function(t) sin(4*pi*t),
function(t) sin(6*pi*t)
),
list(
function(t) cos(3*pi*t),
function(t) cos(pi*t),
function(t) cos(5*pi*t)
)
)
lambdas = c(5, 3, 1)
X = FunOnFun::simMFPCA(16, t, n, 3, mean_funs, eigen_funs_list, lambdas, response = FALSE)Response
mean_funs = list(
function(t) 6*exp(-(t-1)^2),
function(t) -2*14^(t-0.5)
)
eigen_funs_list = list(
list(
function(t) cos(9*pi*t),
function(t) cos(5*pi*t),
function(t) cos(2*pi*t)
),
list(
function(t) sin(3*pi*t),
function(t) sin(5*pi*t),
function(t) sin(7*pi*t)
)
)
B = matrix(c(-1, 0.5, 0.1, -0.5, 1, 0.1, 0.5, 0.5, -0.05),
nrow = components,
ncol = components)
# B = matrix(c(2, 1, 2, -1, -4, 1, 1, 3, 1),
# nrow = components,
# ncol = components)
# B = diag(c(3, 3, 3))
Y = FunOnFun::simMFPCA(16, t, n, 3, mean_funs, eigen_funs_list, lambdas, response = TRUE, B = B)
set.seed(0)
sigma = 0.001
E = matrix(rnorm(2*length(t)*n, mean = 0, sd = sigma), n, 2*length(t))
Y$X = Y$X + EOrthogonalize Response
# cov_Y = Y$phi %*% t(B) %*% t(Y$D) %*% Y$D %*% B %*% t(Y$phi) + diag(rep(sigma^2, 2*length(t)))
popEig = FunOnFun::populationEigen(Y, B, t, sigma)
par(mfrow=c(1, 2))
image(popEig$pop_covY)
image(cov(Y$X))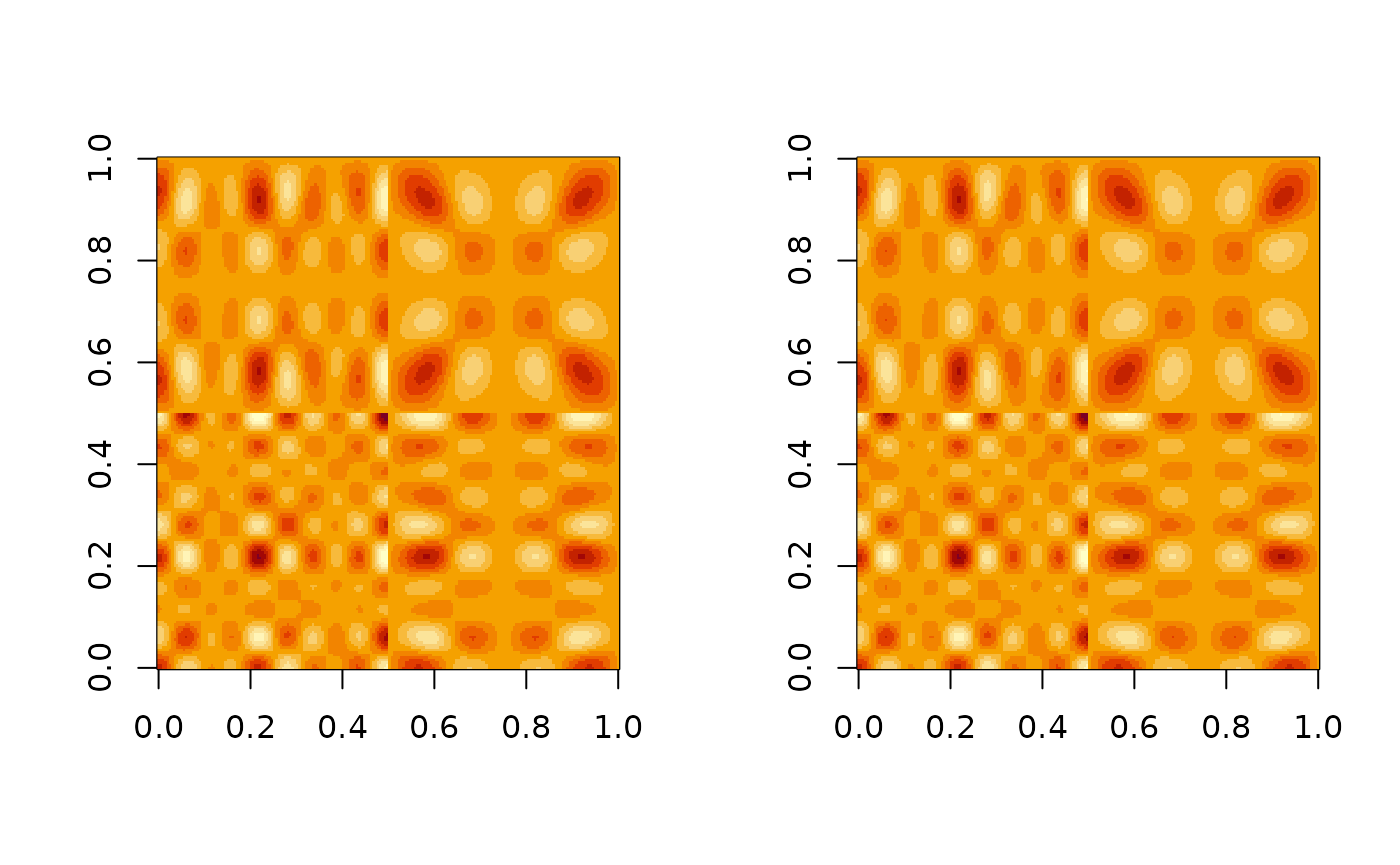
Population Eigenfunctions (Orthogonalized)
# TODO: Make this into a function too; get phi_Y_df, Y_xi_pop, and B_PC_PC
# TODO: Create a FunOnFun regression function that returns B;
# TODO: Make simMFPCA return a class
# phi_Y_df = (eigen(cov_Y)$vectors[, 1:3]*sqrt(2*length(t))) %>% as.data.frame()
popEig$pop_phiY
# Y_xi_pop = Y$X %*% (eigen(cov_Y)$vectors[, 1:3])/sqrt(2*length(t))
popEig$pop_xiY
# B_PC_PC = B %*% t(Y$phi) %*% (eigen(cov_Y)$vectors[, 1:3])/sqrt(2*length(t))
popEig$pop_B
# t(B_PC_PC) %*% Y$D %*% Y$D %*% B_PC_PC
t(popEig$pop_B) %*% Y$D %*% Y$D %*% popEig$pop_BPlot Simulated Data
matplot(t(X$X),
type='l',
ylab='X(t)',
xlab='time',
main='Plot of predictor curves',
col=rgb(0,0,0,alpha=0.4))
matlines(apply(t(X$X), 1, mean),
type='l',
lwd=3,
lty=1,
col="red")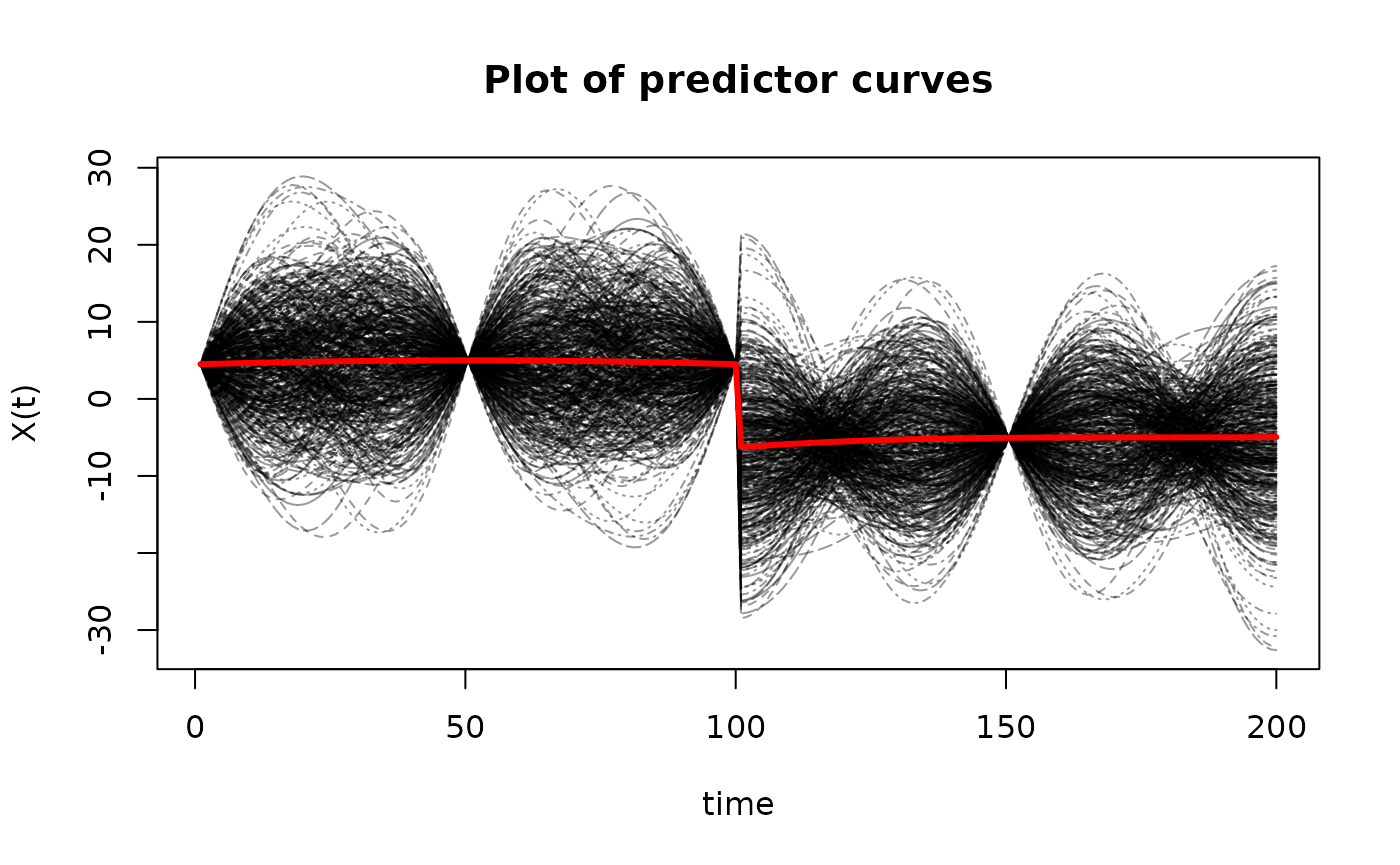
matplot(t(Y$X),
type='l',
ylab='Y(t)',
xlab='time',
main='Plot of response curves',
col=rgb(0,0,0,alpha=0.6))
matlines(apply(t(Y$X), 1, mean),
type='l',
lwd=3,
lty=1,
col = "red")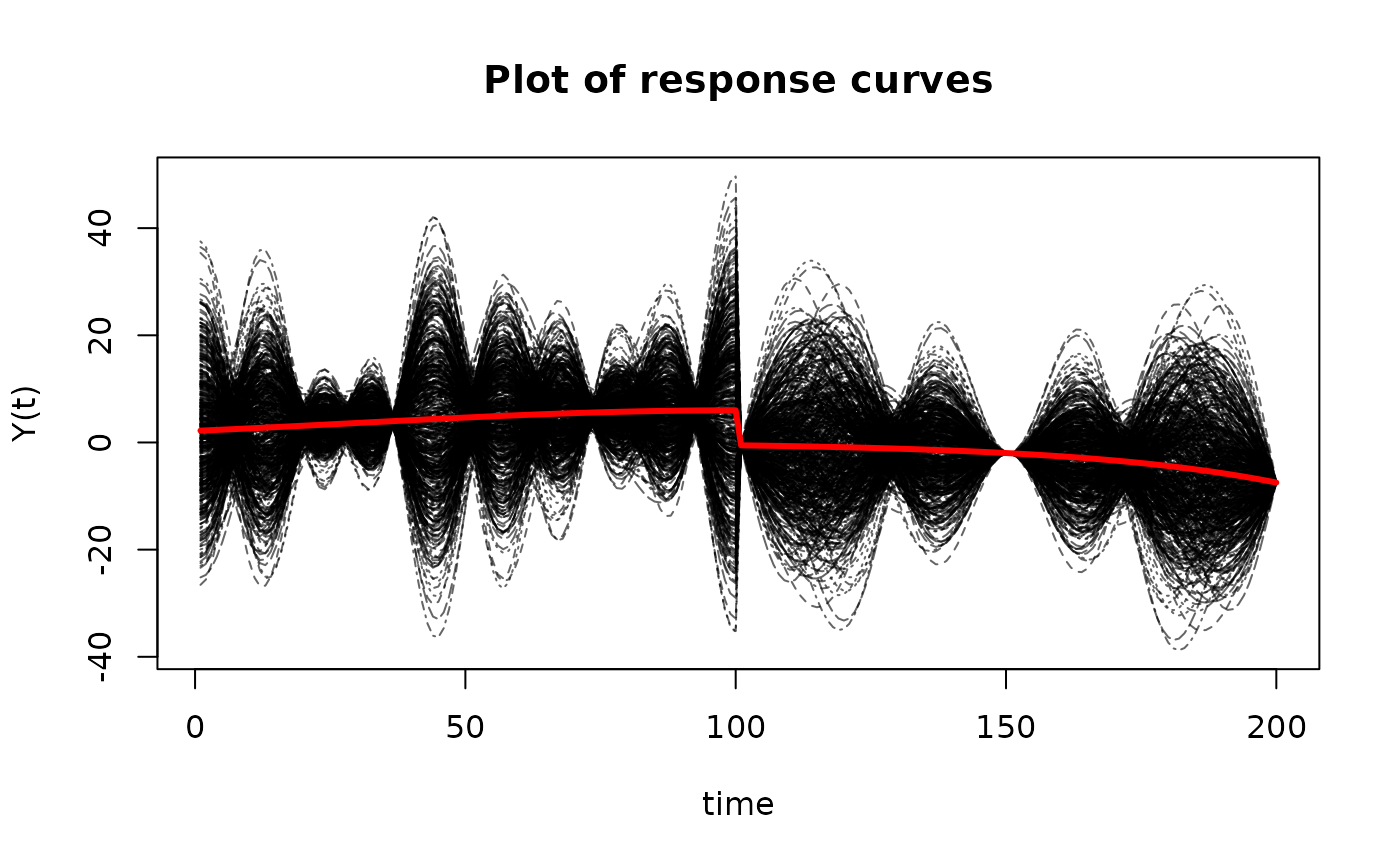
FPCA
df = X_miss %>% FunOnFun::tibbleFormat(t) %>% FunOnFun::fpcaFormat(id_col = "id")
#> Registered S3 method overwritten by 'tsibble':
#> method from
#> as_tibble.grouped_df dplyr
df_Y = Y_miss %>% FunOnFun::tibbleFormat(t) %>% FunOnFun::fpcaFormat(id_col = "id")
res_X1 = fdapace::FPCA(df$Variable1,
df$Time,
list(dataType = "Sparse",
error = F,
kernel = "epan",
verbose = F,
nRegGrid = length(t)))
res_X2 = fdapace::FPCA(df$Variable2,
df$Time,
list(dataType = "Sparse",
error = F,
kernel = "epan",
verbose = F,
nRegGrid = length(t)))
res_Y1 = fdapace::FPCA(df_Y$Variable1,
df_Y$Time,
list(dataType = "Sparse",
error = T,
kernel = "epan",
verbose = F,
nRegGrid = length(t),
methodSelectK = 3))
res_Y2 = fdapace::FPCA(df_Y$Variable2,
df_Y$Time,
list(dataType = "Sparse",
error = T,
kernel = "epan",
verbose = F,
nRegGrid = length(t),
methodSelectK = 3))Visualize FPCA
act = data.frame(act1 = Y$mu[1:100],
act2 = Y$mu[101:200])
hat = data.frame(hat1 = res_Y1$mu,
hat2 = res_Y2$mu)
hat %>%
ggplot() +
geom_line(aes(x = seq(0, 1, length.out = 100), y = hat1)) +
geom_line(data = act, aes(x = seq(0, 1, length.out = 100), y = act1), linetype = "dashed") +
theme_bw()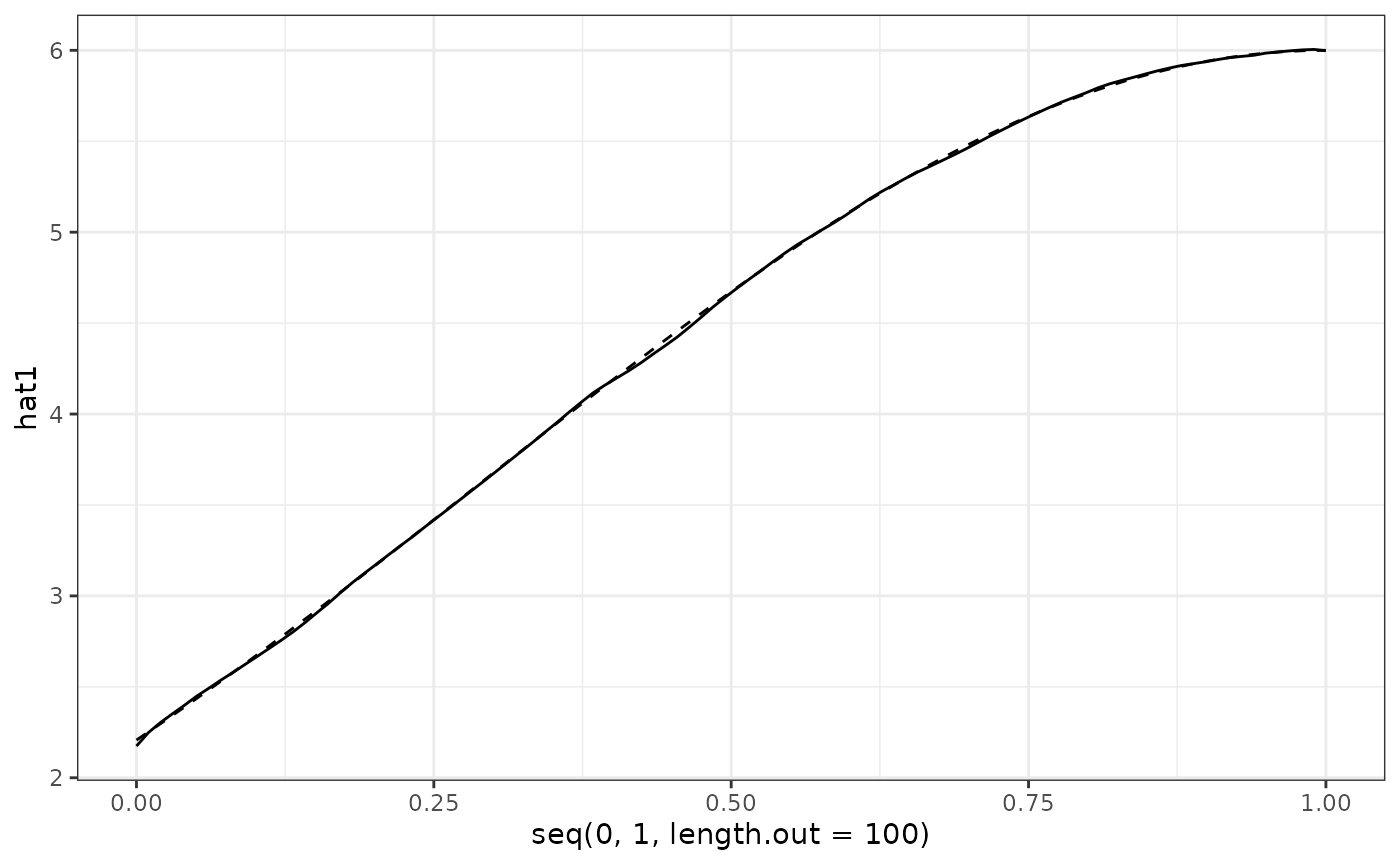
hat %>%
ggplot() +
geom_line(aes(x = seq(0, 1, length.out = 100), y = hat2)) +
geom_line(data = act, aes(x = seq(0, 1, length.out = 100), y = act2), linetype = "dashed") +
theme_bw()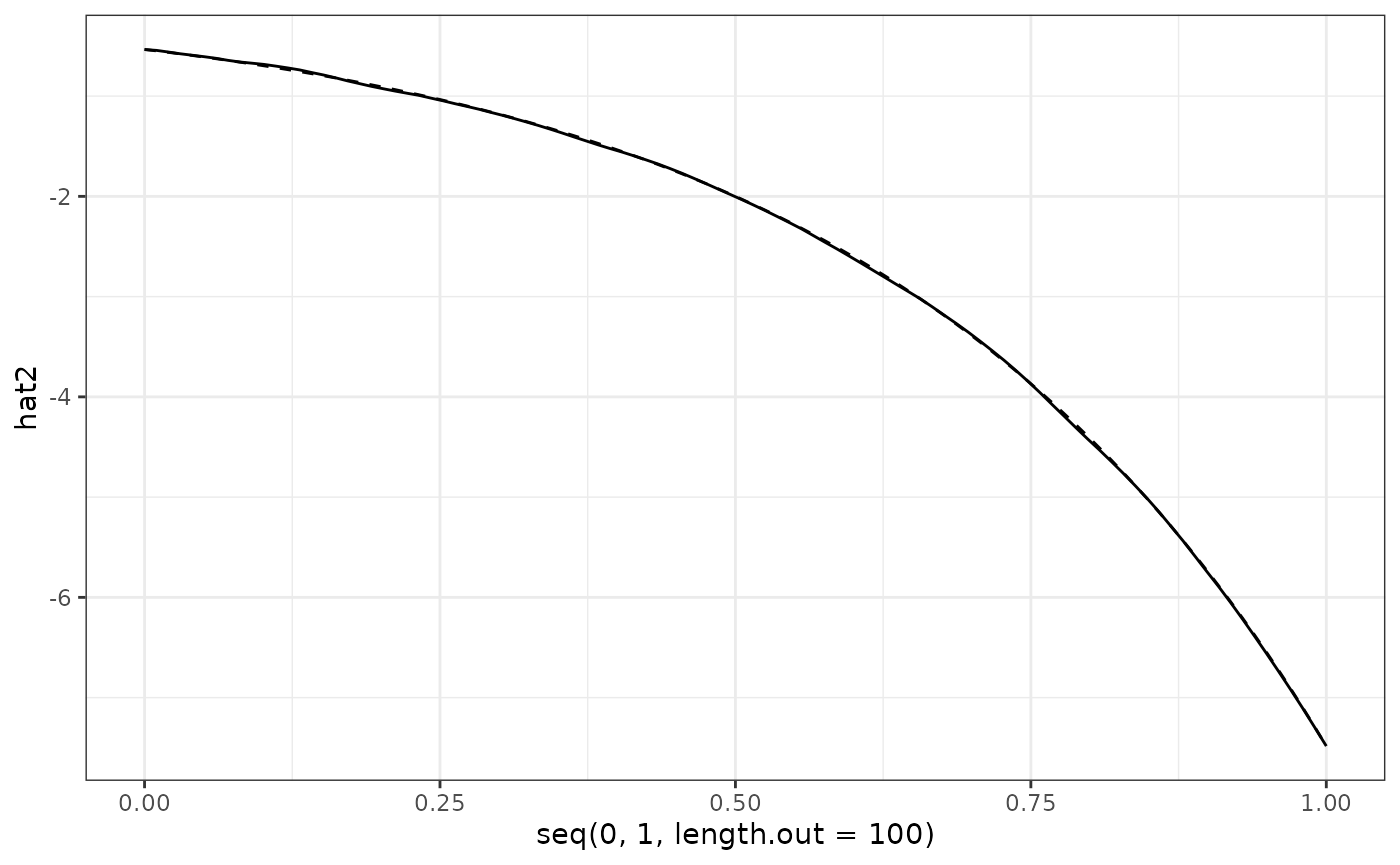
#phi_X2_df = Y$phi[101:200,] %>% as.data.frame()
phi_Y1_df = (eigen(popEig$pop_covY[1:100,1:100])$vectors[, 1:3]*sqrt(100)) %>% as.data.frame()
res_Y1$phi[, 1:3] %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = t, y = V1), col = "red") +
geom_line(data = phi_Y1_df, aes(x = t, y = V1), col = "red", linetype = "dashed") +
theme_bw()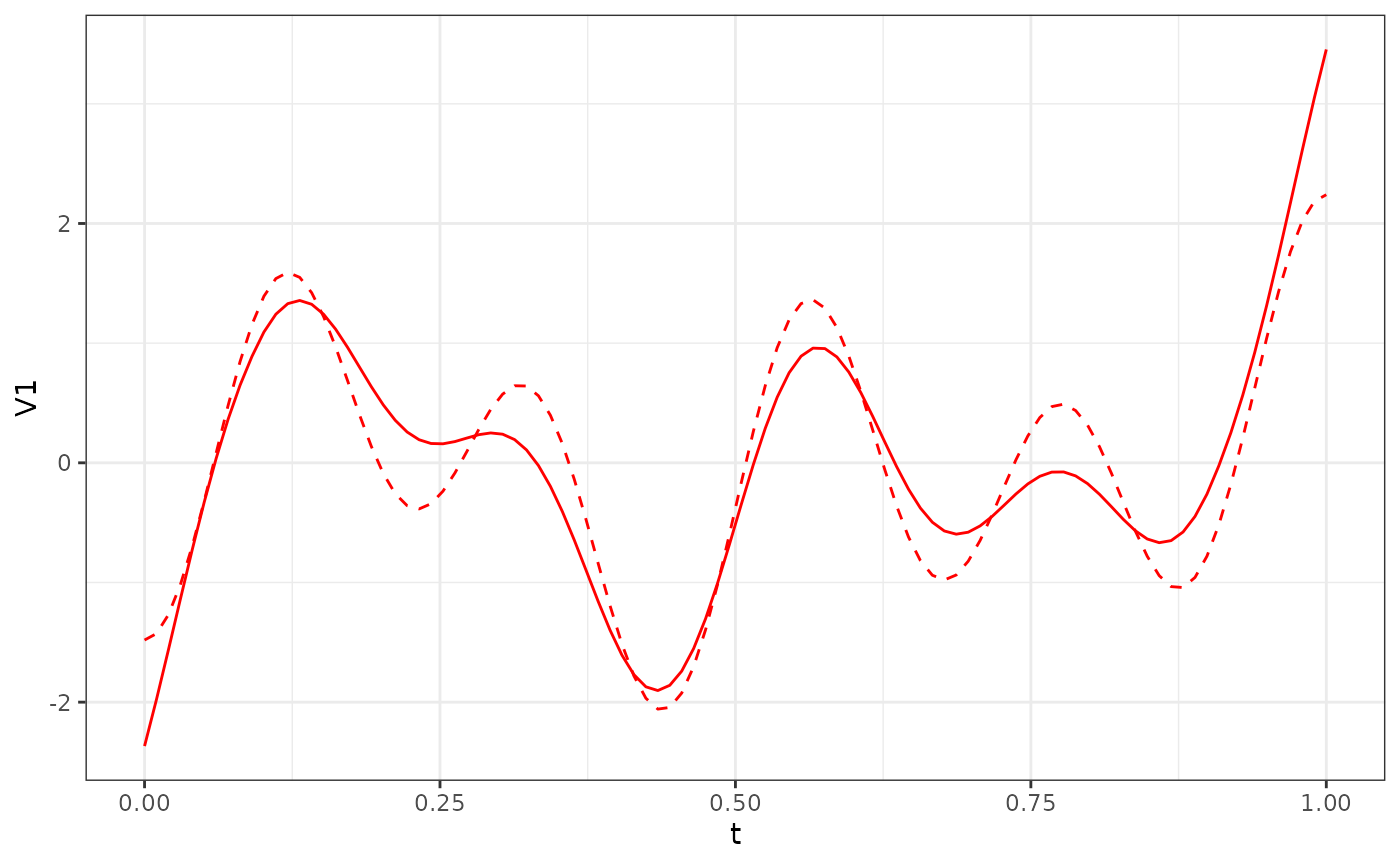
res_Y1$phi[, 1:3] %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = t, y = V2), col = "red") +
geom_line(data = phi_Y1_df, aes(x = t, y = V2), col = "red", linetype = "dashed") +
theme_bw()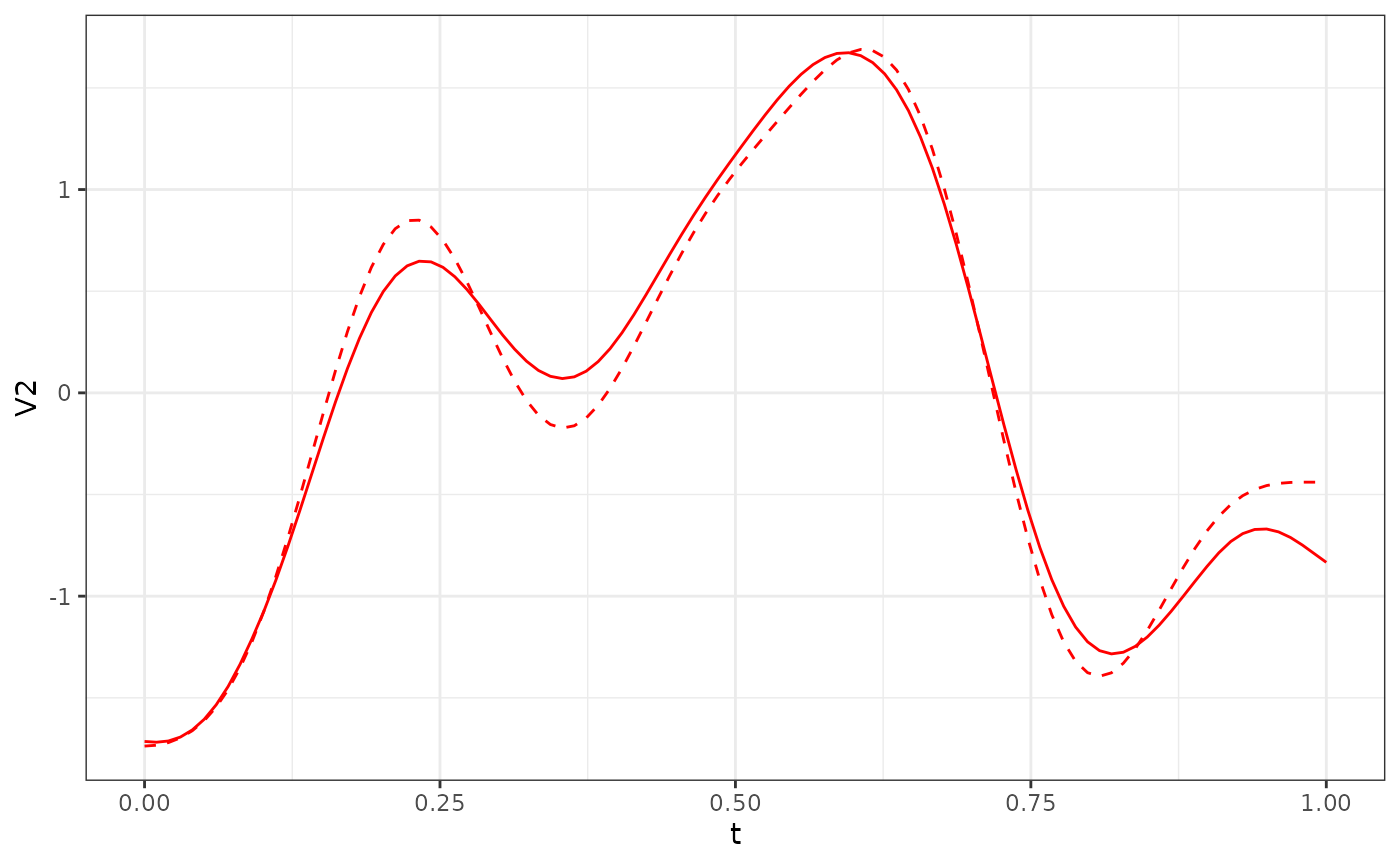
res_Y1$phi[, 1:3] %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = t, y = -V3), col = "red") +
geom_line(data = phi_Y1_df, aes(x = t, y = V3), col = "red", linetype = "dashed") +
theme_bw()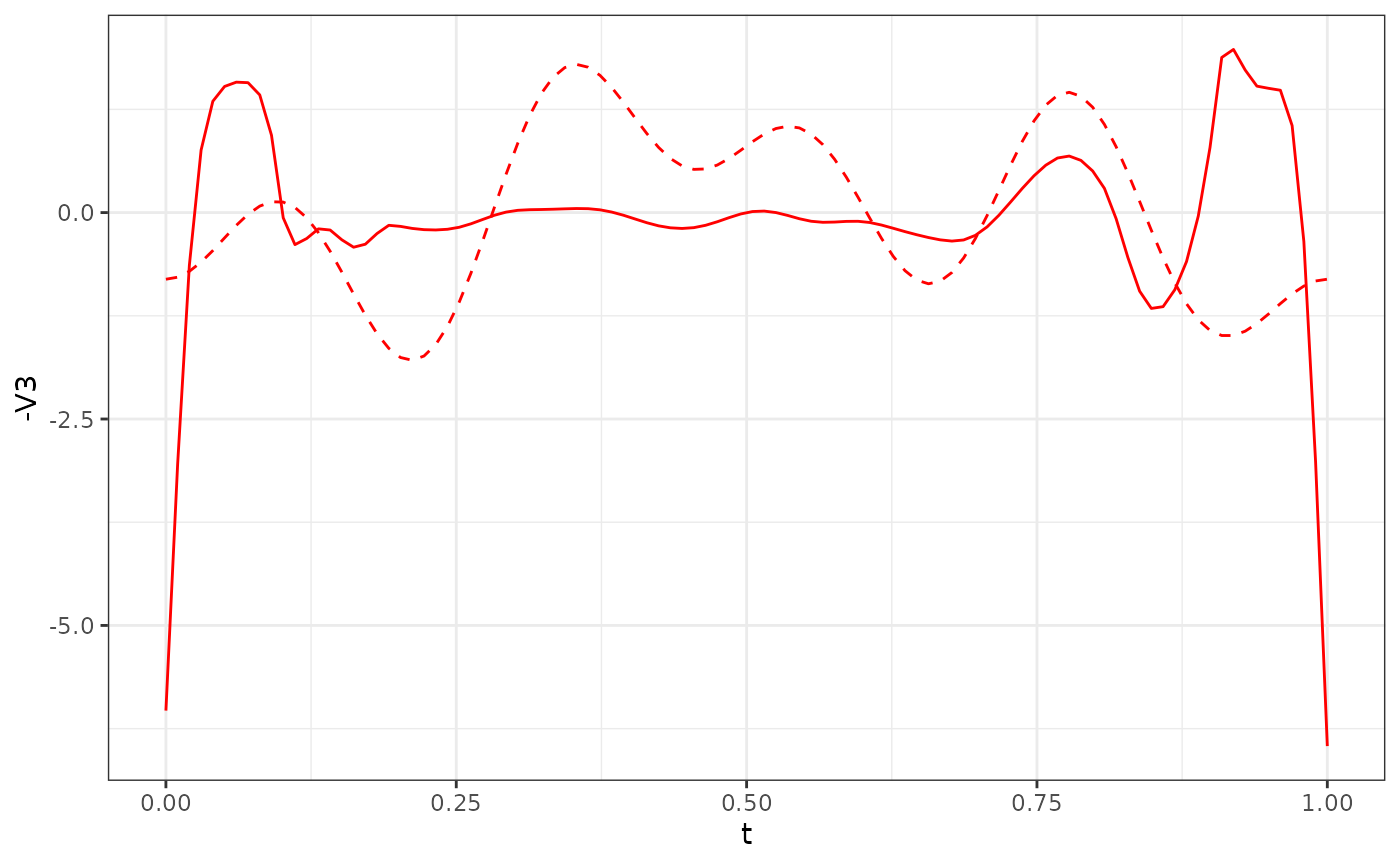
phi_Y2_df = (eigen(popEig$pop_covY[101:200,101:200])$vectors[, 1:3]*sqrt(length(t))) %>% as.data.frame()
res_Y2$phi[, 1:3] %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = t, y = V1), col = "red") +
geom_line(data = phi_Y2_df, aes(x = t, y = V1), col = "red", linetype = "dashed") +
theme_bw()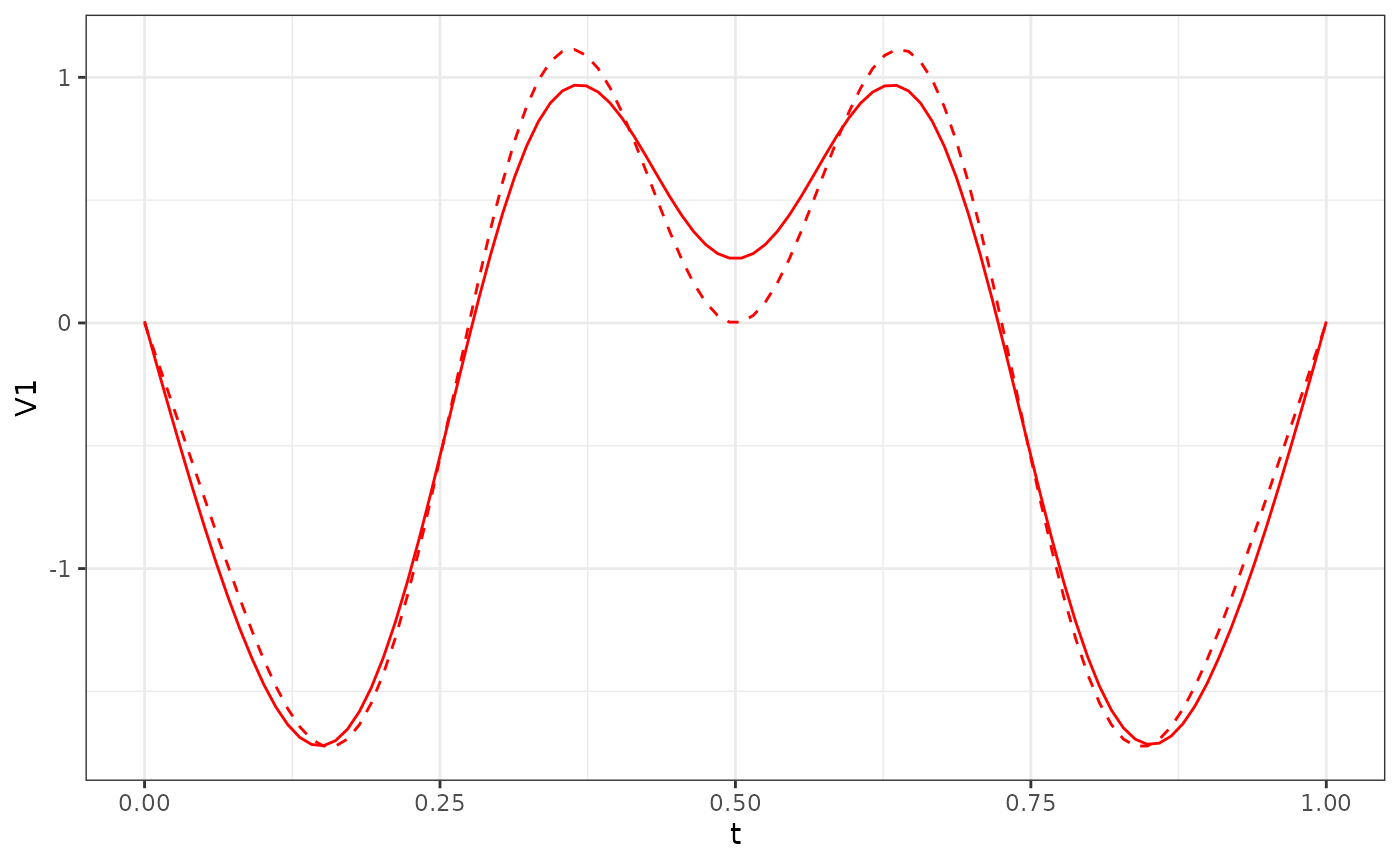
res_Y2$phi[, 1:3] %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = t, y = V2), col = "red") +
geom_line(data = phi_Y2_df, aes(x = t, y = V2), col = "red", linetype = "dashed") +
theme_bw()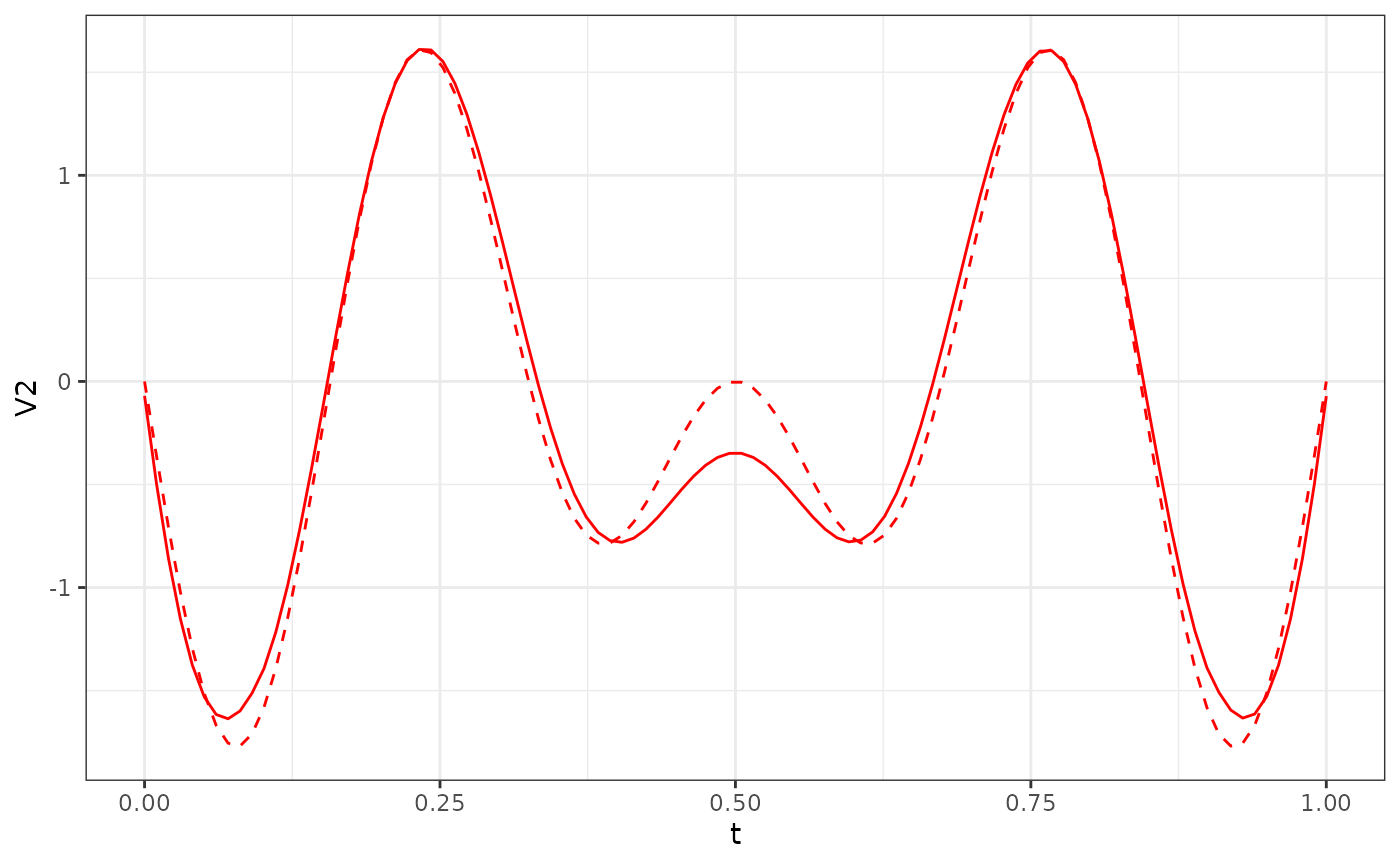
res_Y2$phi[, 1:3] %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = t, y = V3), col = "red") +
geom_line(data = phi_Y2_df, aes(x = t, y = V3), col = "red", linetype = "dashed") +
theme_bw()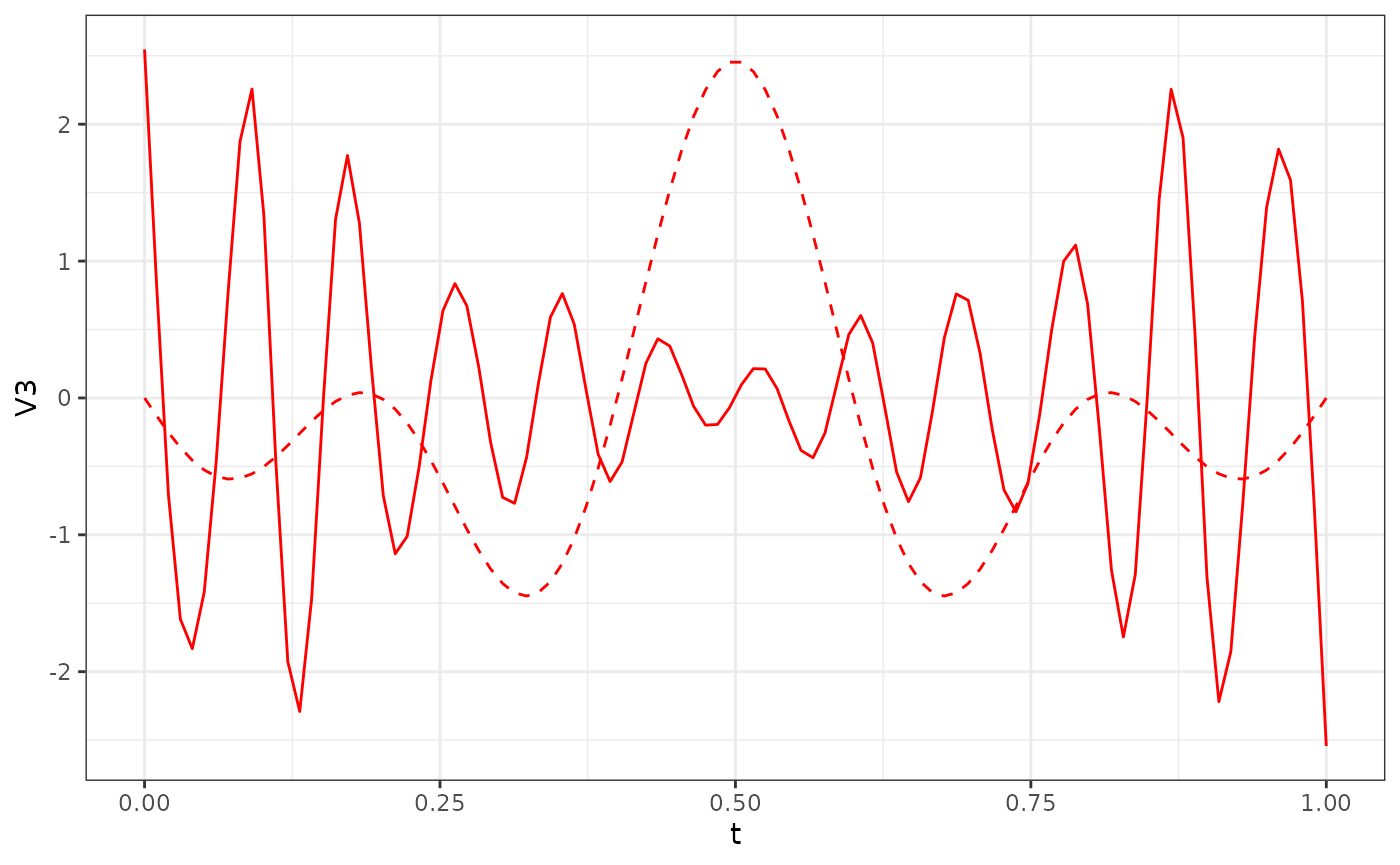
Irregular MFPCA
res = FunOnFun::irregMFPCA(components = 3,
split = T,
res_X1,
res_X2)
res_Y = FunOnFun::irregMFPCA(components = 3,
split = T,
res_Y1,
res_Y2)Check results
eigenf = res$unstacked_phi
colnames(eigenf) = c("var_1_1", "var_1_2", "var_1_3", "var_2_1", "var_2_2", "var_2_3")
eigens = res$xi
colnames(eigens) = c("comp_1", "comp_2", "comp_3")
eigenf %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = seq(0, 1, length.out = 100), y = var_1_1), col = "red") +
geom_line(aes(x = seq(0, 1, length.out = 100), y = X$phi[1:100, 1]), col = "black", linetype = "dotted") +
theme_bw()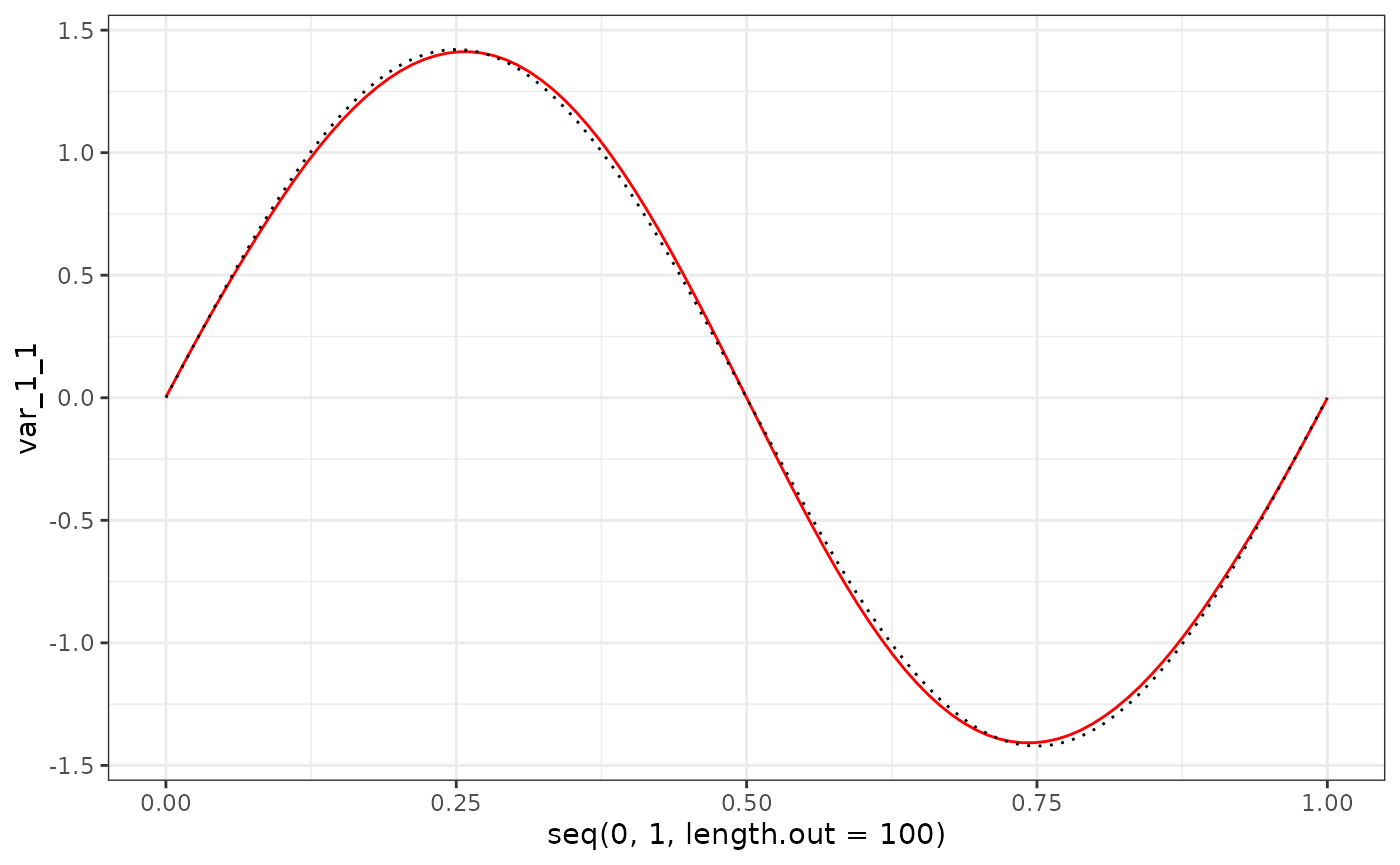
eigenf %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = seq(0, 1, length.out = 100), y = -var_1_2), col = "red") +
geom_line(aes(x = seq(0, 1, length.out = 100), y = X$phi[1:100, 2]), col = "black", linetype = "dotted") +
theme_bw()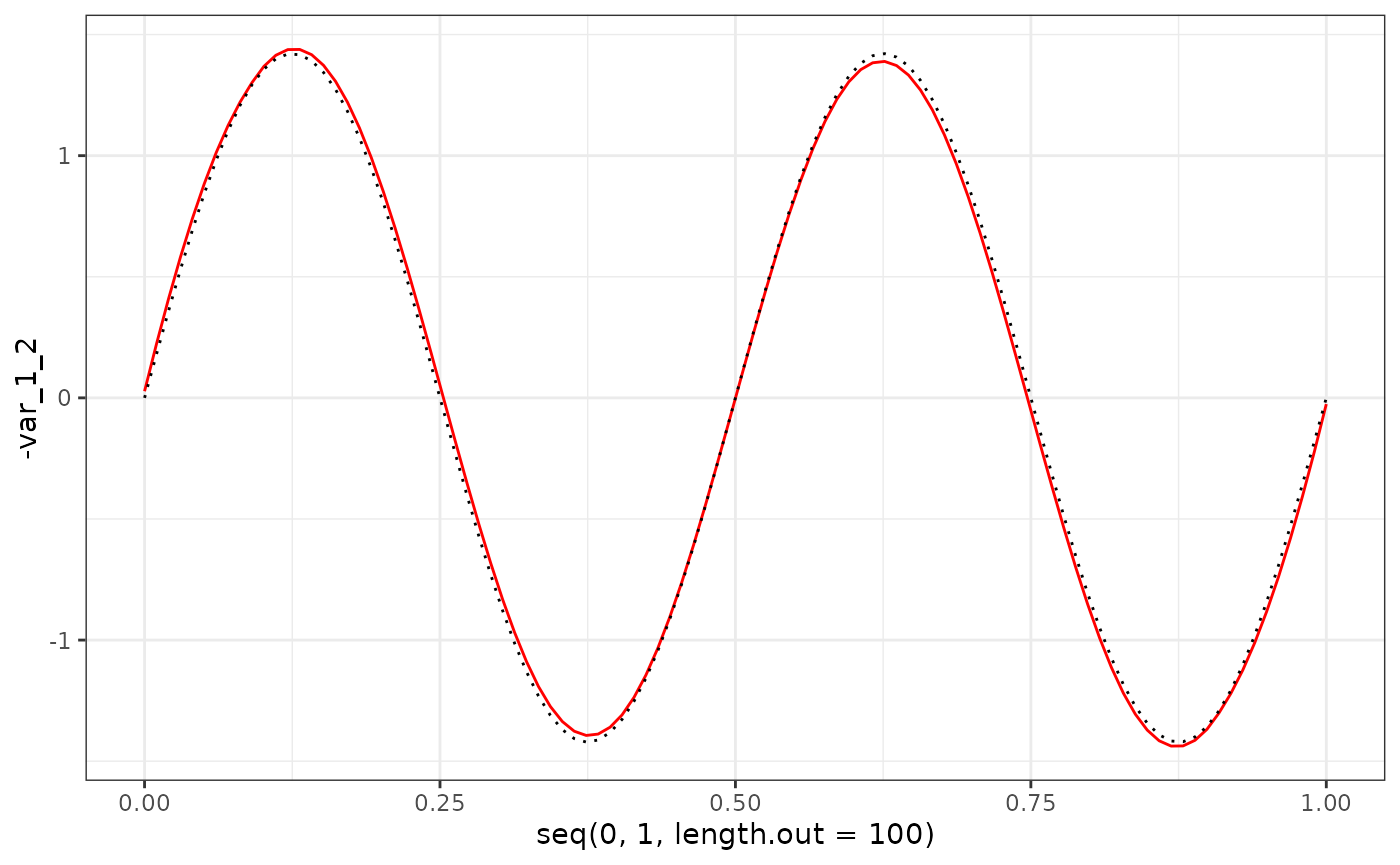
eigenf %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = seq(0, 1, length.out = 100), y = -var_1_3), col = "red") +
geom_line(aes(x = seq(0, 1, length.out = 100), y = X$phi[1:100, 3]), col = "black", linetype = "dotted") +
theme_bw()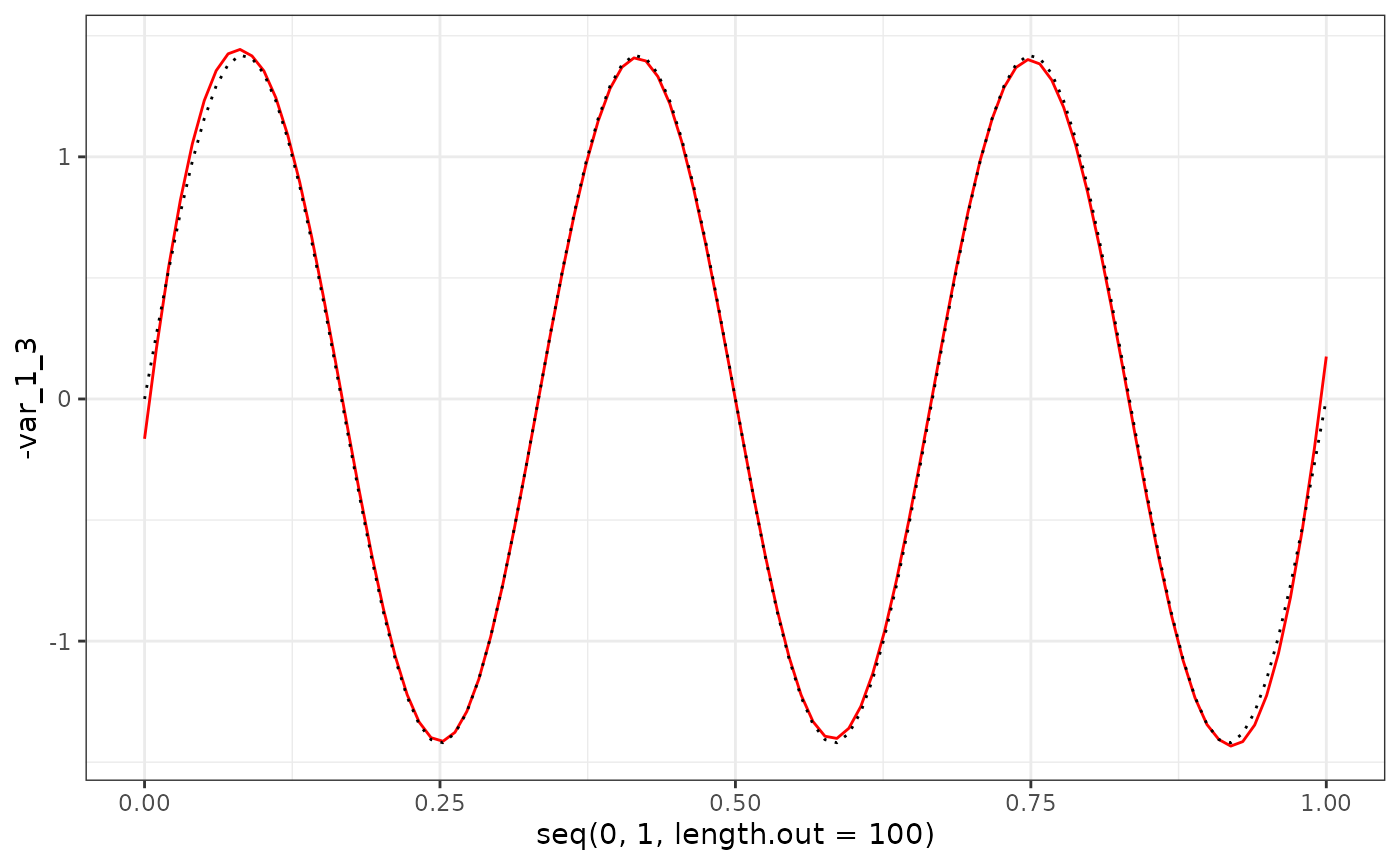
eigenf %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = seq(0, 1, length.out = 100), y = var_2_1), col = "red") +
geom_line(aes(x = seq(0, 1, length.out = 100), y = X$phi[101:200, 1]), col = "black", linetype = "dotted") +
theme_bw()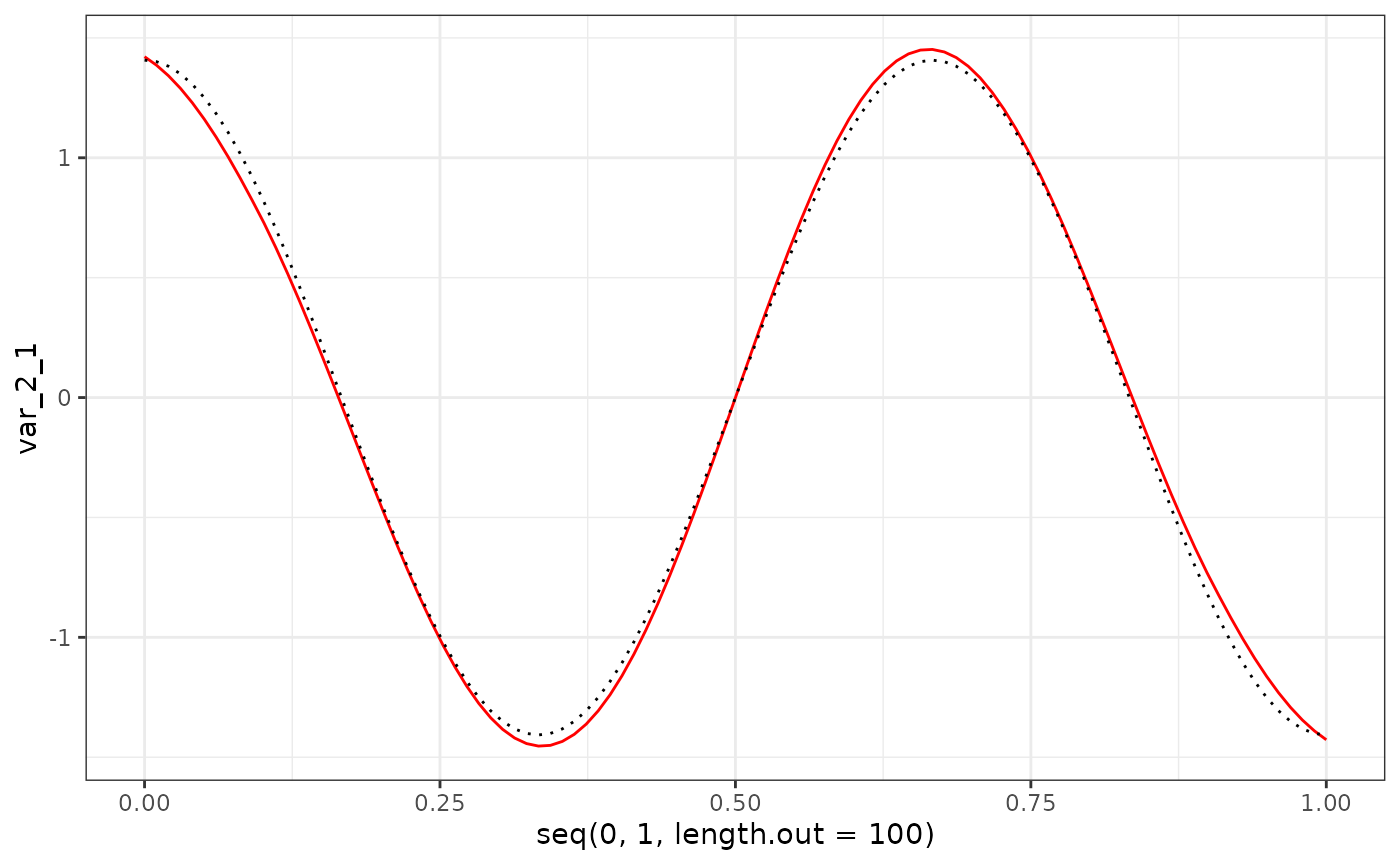
eigenf %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = seq(0, 1, length.out = 100), y = -var_2_2), col = "red") +
geom_line(aes(x = seq(0, 1, length.out = 100), y = X$phi[101:200, 2]), col = "black", linetype = "dotted") +
theme_bw()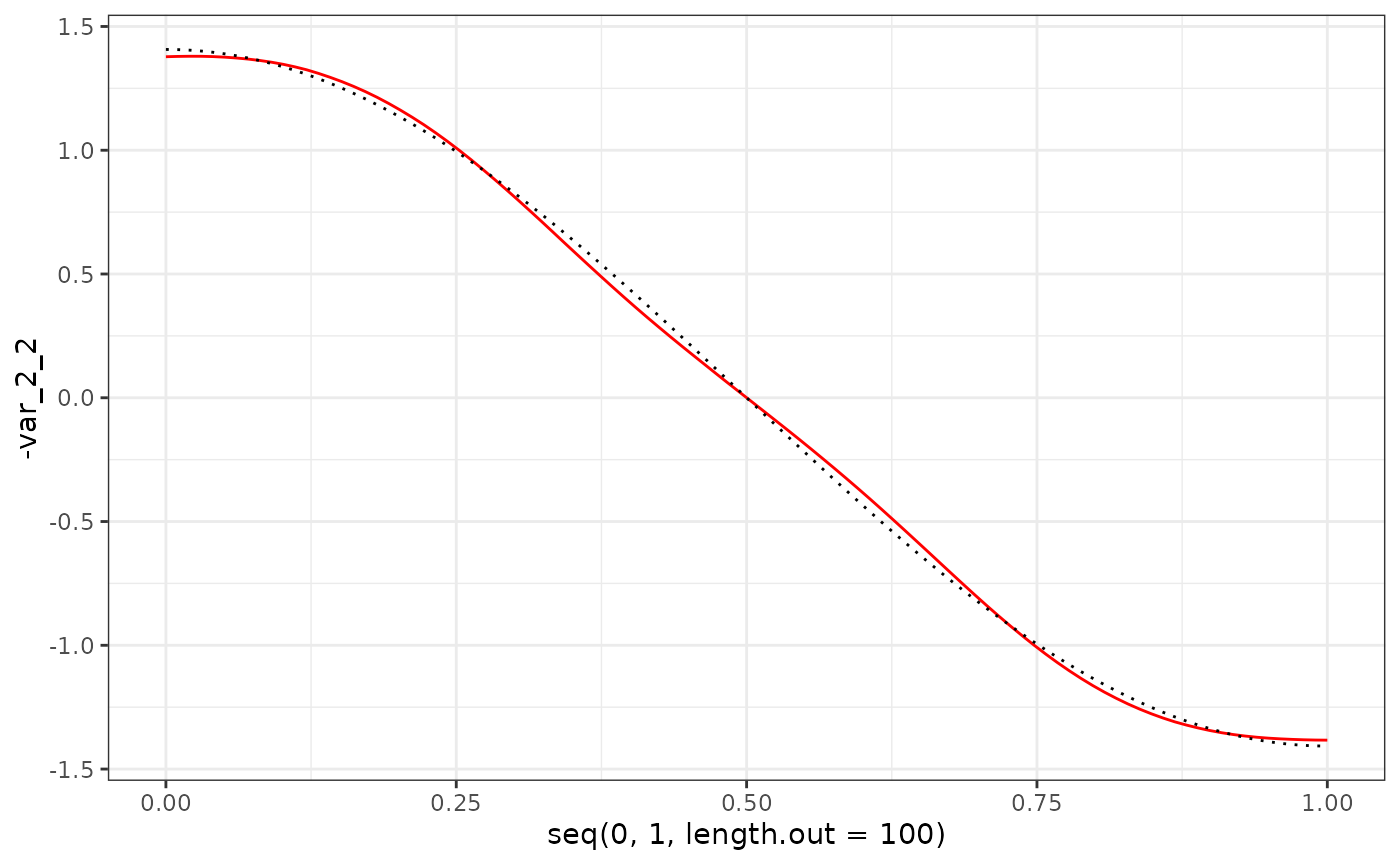
eigenf %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = seq(0, 1, length.out = 100), y = -var_2_3), col = "red") +
geom_line(aes(x = seq(0, 1, length.out = 100), y = X$phi[101:200, 3]), col = "black", linetype = "dotted") +
theme_bw()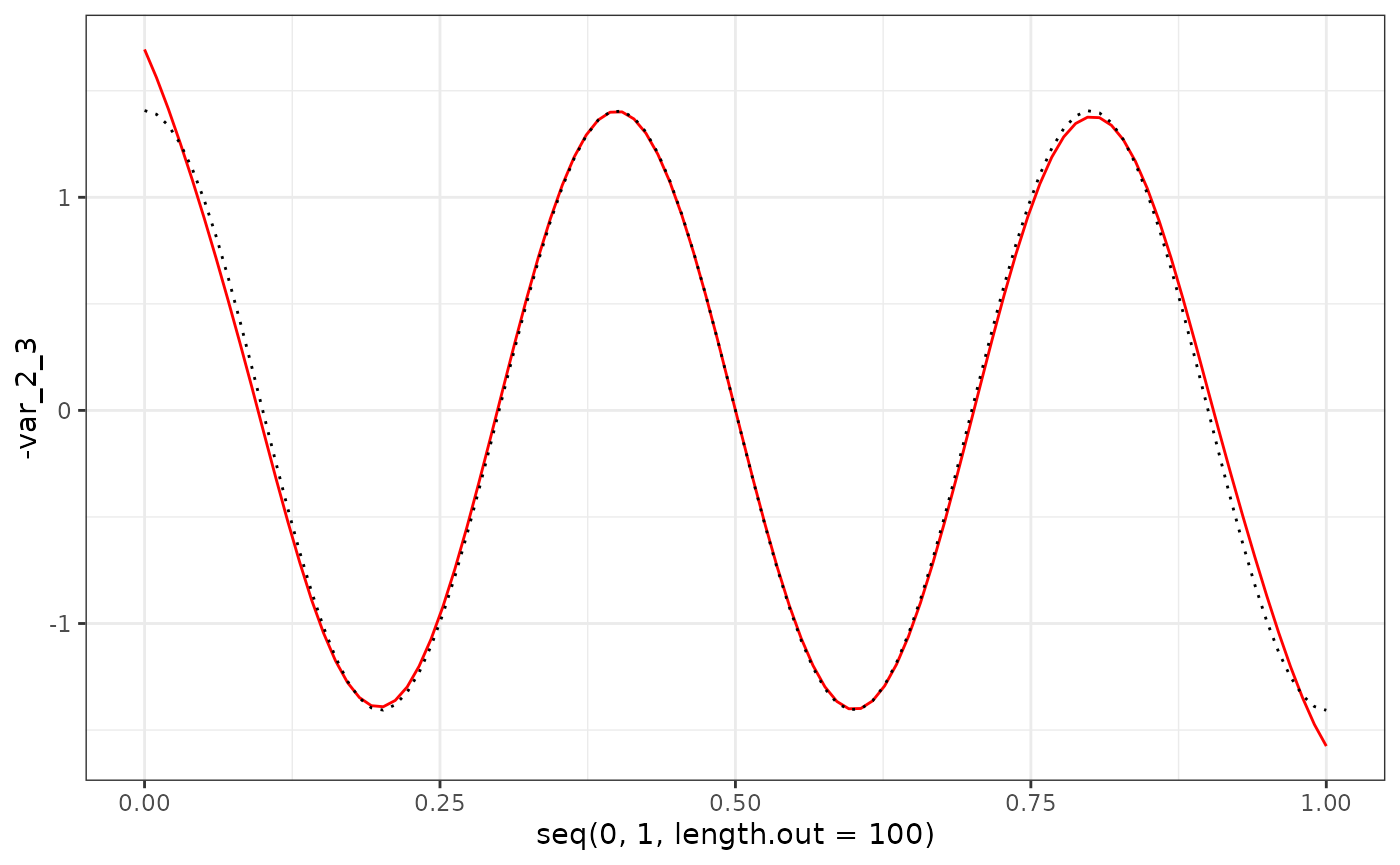
eigens = data.frame(est1 = eigens[,1]/sqrt(res$Dhat[1,1]),
est2 = eigens[,2]/sqrt(res$Dhat[2,2]),
est3 = eigens[,3]/sqrt(res$Dhat[3,3]),
act1 = X$xi[, 1],
act2 = X$xi[, 2],
act3 = X$xi[, 3])
eigens %>%
ggplot(aes(x = est1, y = act1)) +
geom_point() +
geom_abline(intercept = 0, slope = 1, col = "red") +
geom_smooth(method = "lm", se = F) +
theme_bw()
#> `geom_smooth()` using formula = 'y ~ x'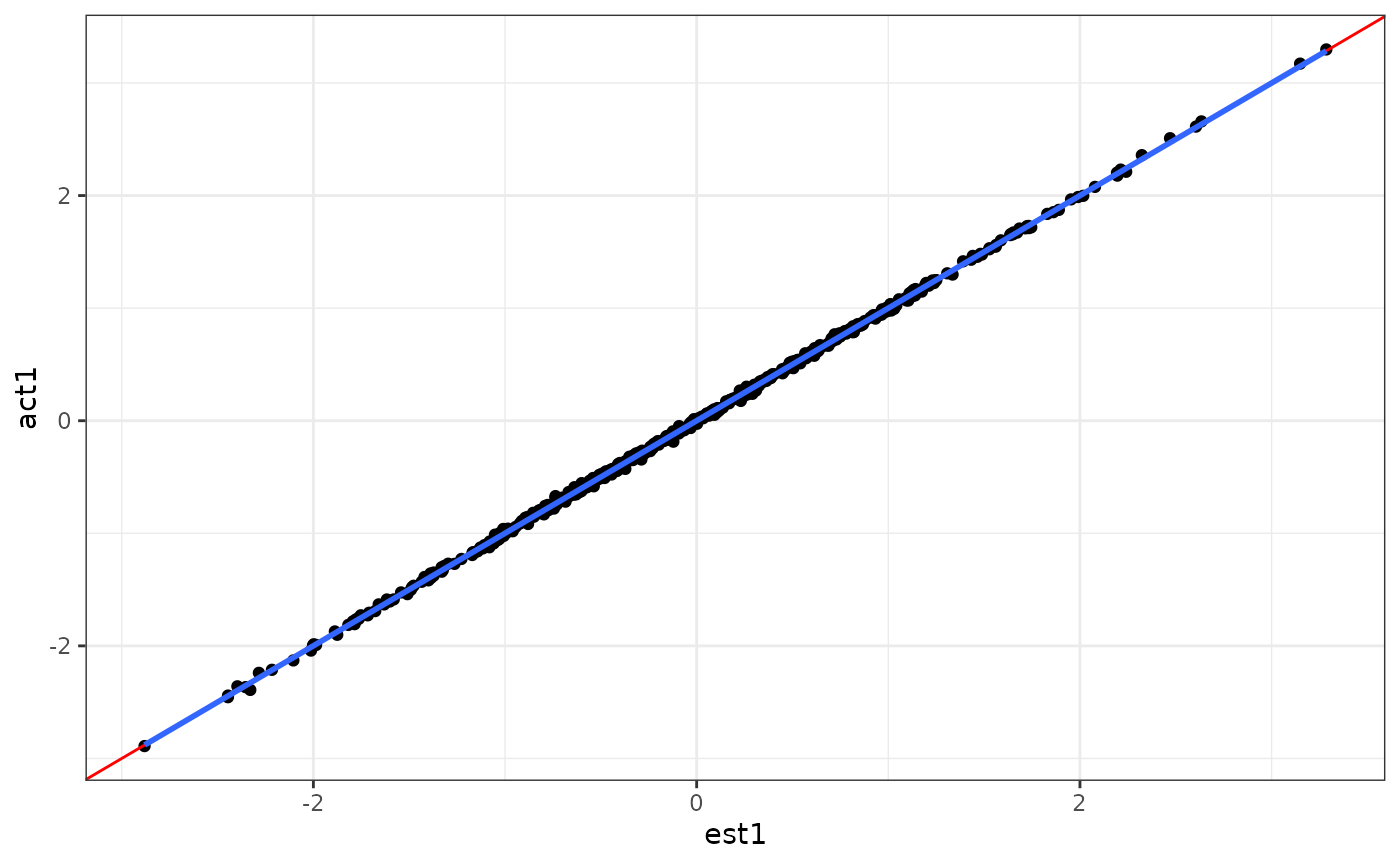
eigens %>%
ggplot(aes(x = -est2, y = act2)) +
geom_point() +
geom_abline(intercept = 0, slope = 1, col = "red") +
geom_smooth(method = "lm", se = F) +
theme_bw()
#> `geom_smooth()` using formula = 'y ~ x'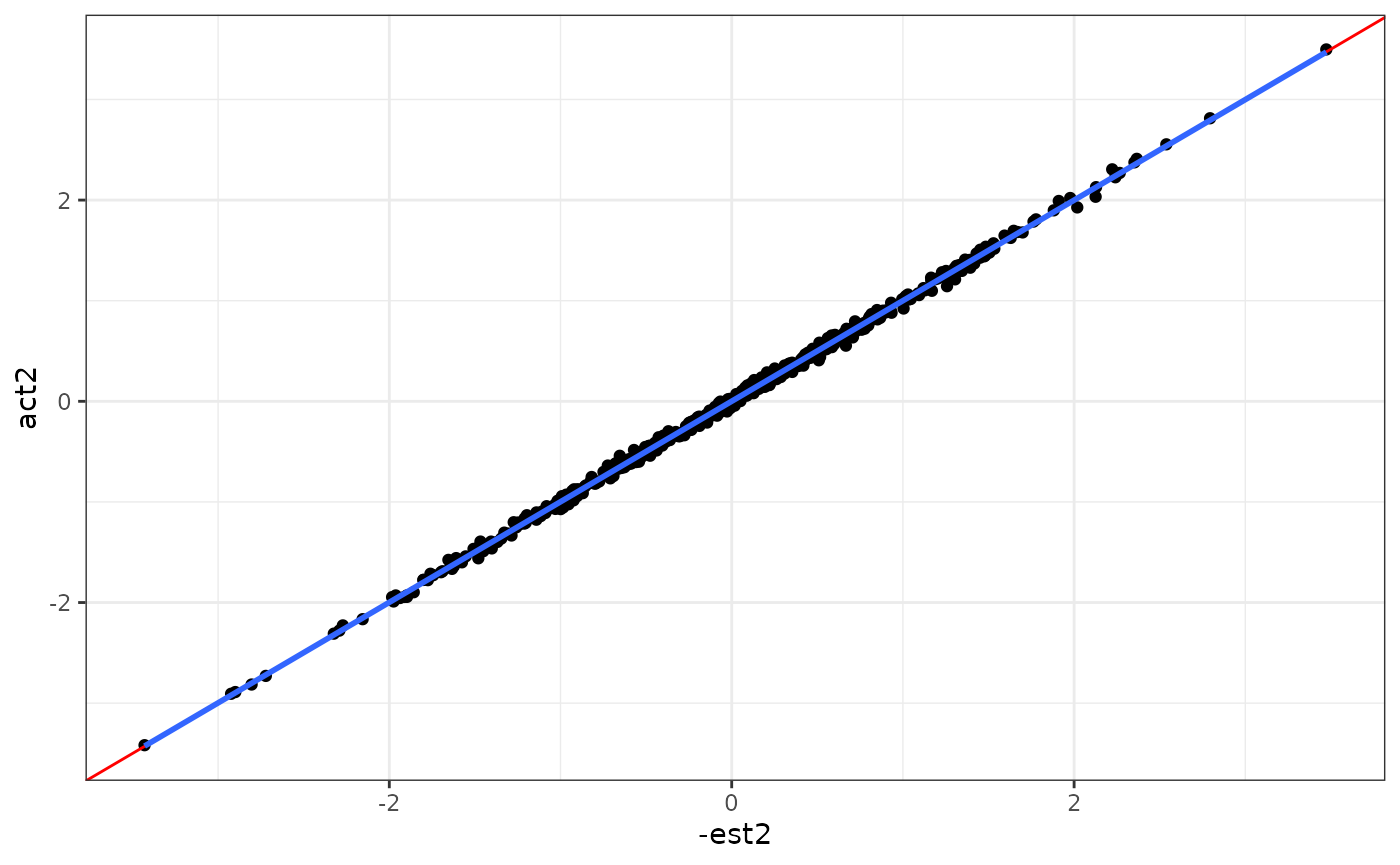
eigens %>%
ggplot(aes(x = -est3, y = act3)) +
geom_point() +
geom_abline(intercept = 0, slope = 1, col = "red") +
geom_smooth(method = "lm", se = F) +
theme_bw()
#> `geom_smooth()` using formula = 'y ~ x'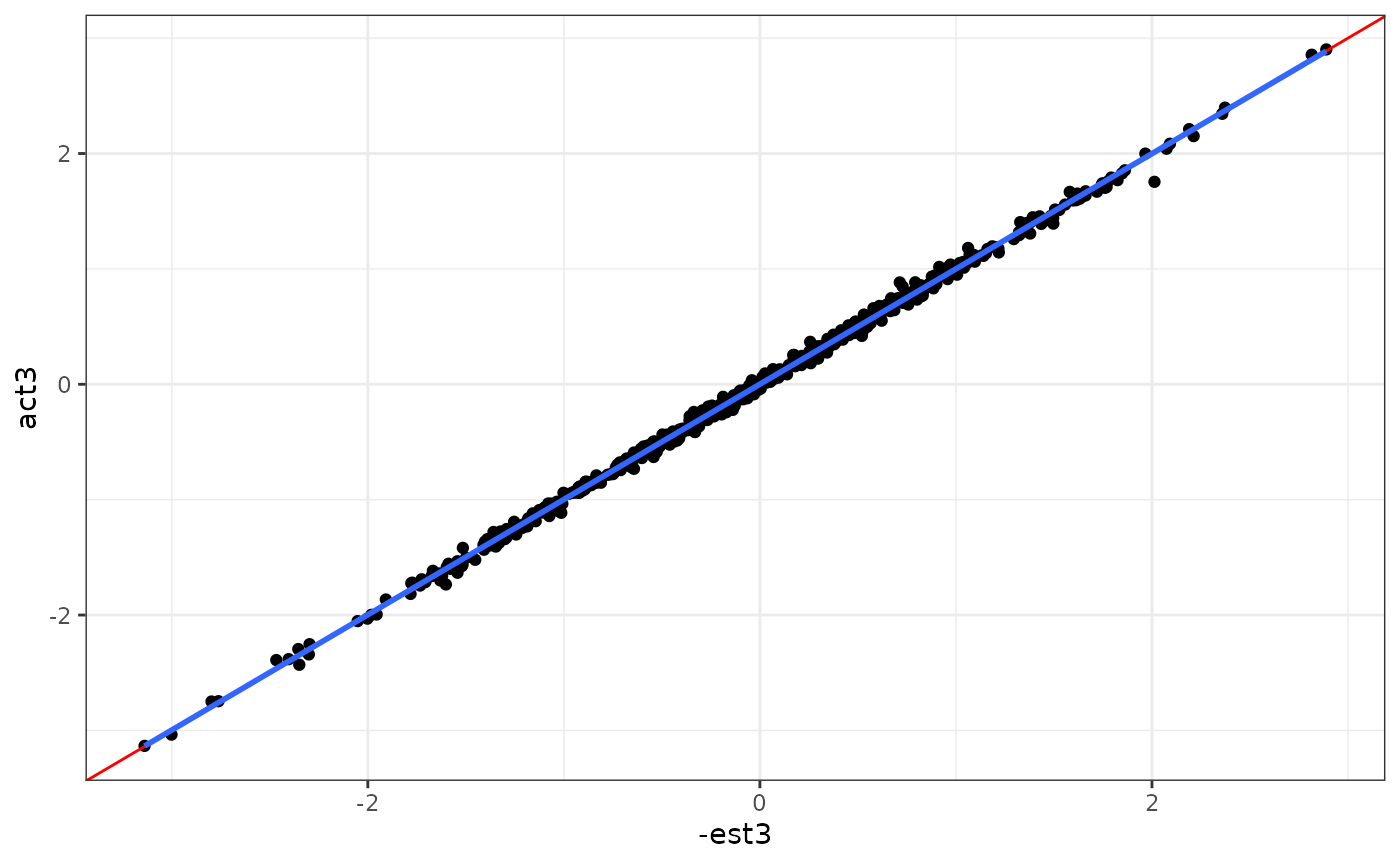
eigenf = res_Y$unstacked_phi
colnames(eigenf) = c("var_1_1", "var_1_2", "var_1_3", "var_2_1", "var_2_2", "var_2_3")
eigens = res_Y$xi %>% sweep(2, sqrt(diag(res_Y$Dhat)), "/")
colnames(eigens) = c("comp_1", "comp_2", "comp_3")
# act = qr.Q(qr(Y$xi %*% B)) * sqrt(n)
# Y_xi_pop = Y$X %*% (eigen(cov_Y)$vectors[, 1:3])/sqrt(n)
# Y_xi_pop = (Y$X - rep(1, n) %*% t(Y$mu)) %*% (eigen(cov_Y)$vectors[, 1:3]) %>% sweep(2, lambdas, "/")/sqrt(n)
#TODO: Figure out how to use normalized scores!! What are the population normalized scores?
# eigens = data.frame(est1 = eigens[,1],
# est2 = eigens[,2],
# est3 = eigens[,3],
# act1 = -act[, 1],
# act2 = act[, 2],
# act3 = -act[, 3])
phi_Y_df = (eigen(popEig$pop_covY)$vectors[, 1:3]*sqrt(2*length(t))) %>% as.data.frame()
eigenf %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = seq(0, 1, length.out = 100), y = var_1_1), col = "red") +
geom_line(aes(x = seq(0, 1, length.out = 100), y = phi_Y_df[1:100, 1]), col = "black", linetype = "dotted") +
theme_bw()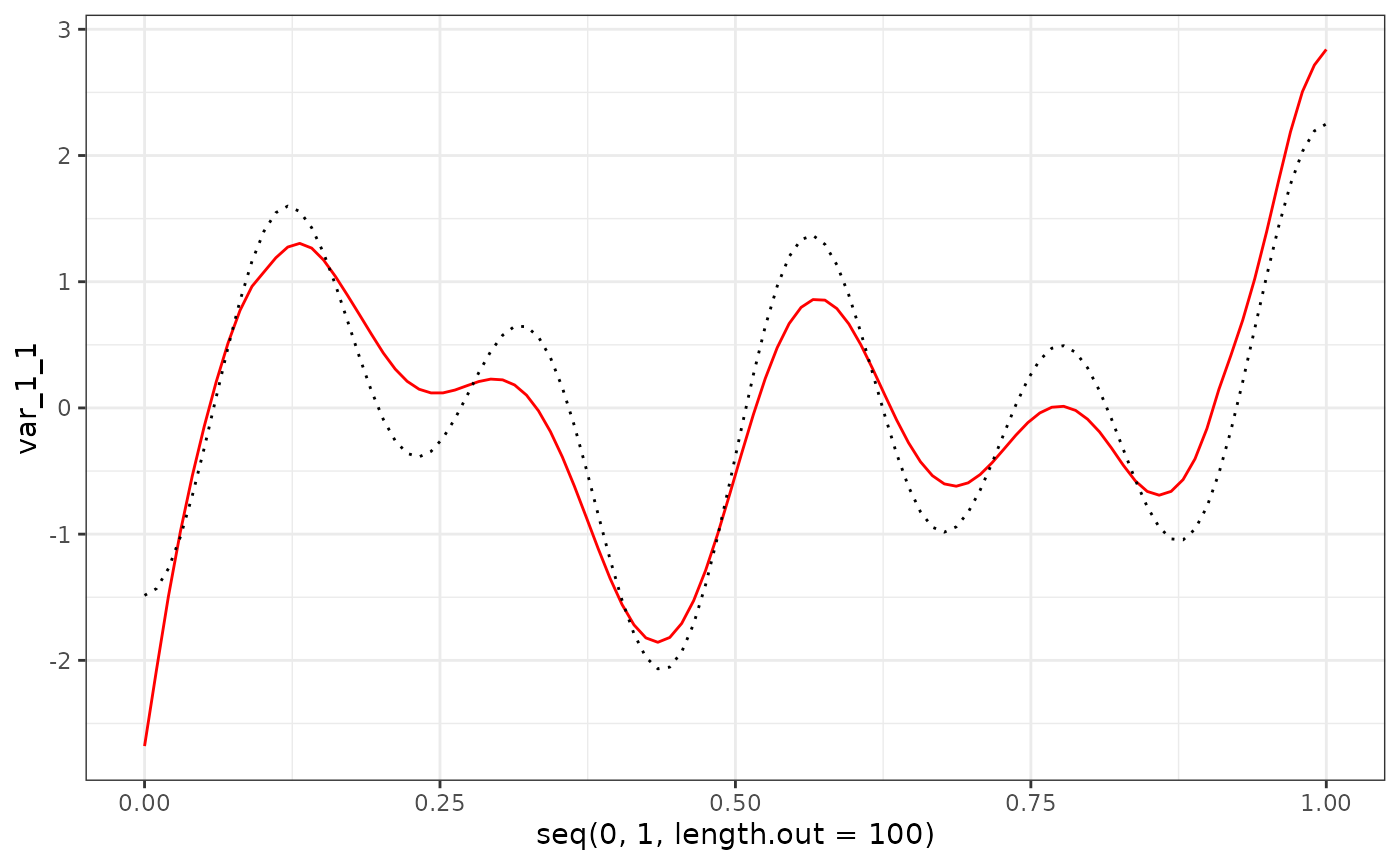
eigenf %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = seq(0, 1, length.out = 100), y = -var_1_2), col = "red") +
geom_line(aes(x = seq(0, 1, length.out = 100), y = phi_Y_df[1:100, 2]), col = "black", linetype = "dotted") +
theme_bw()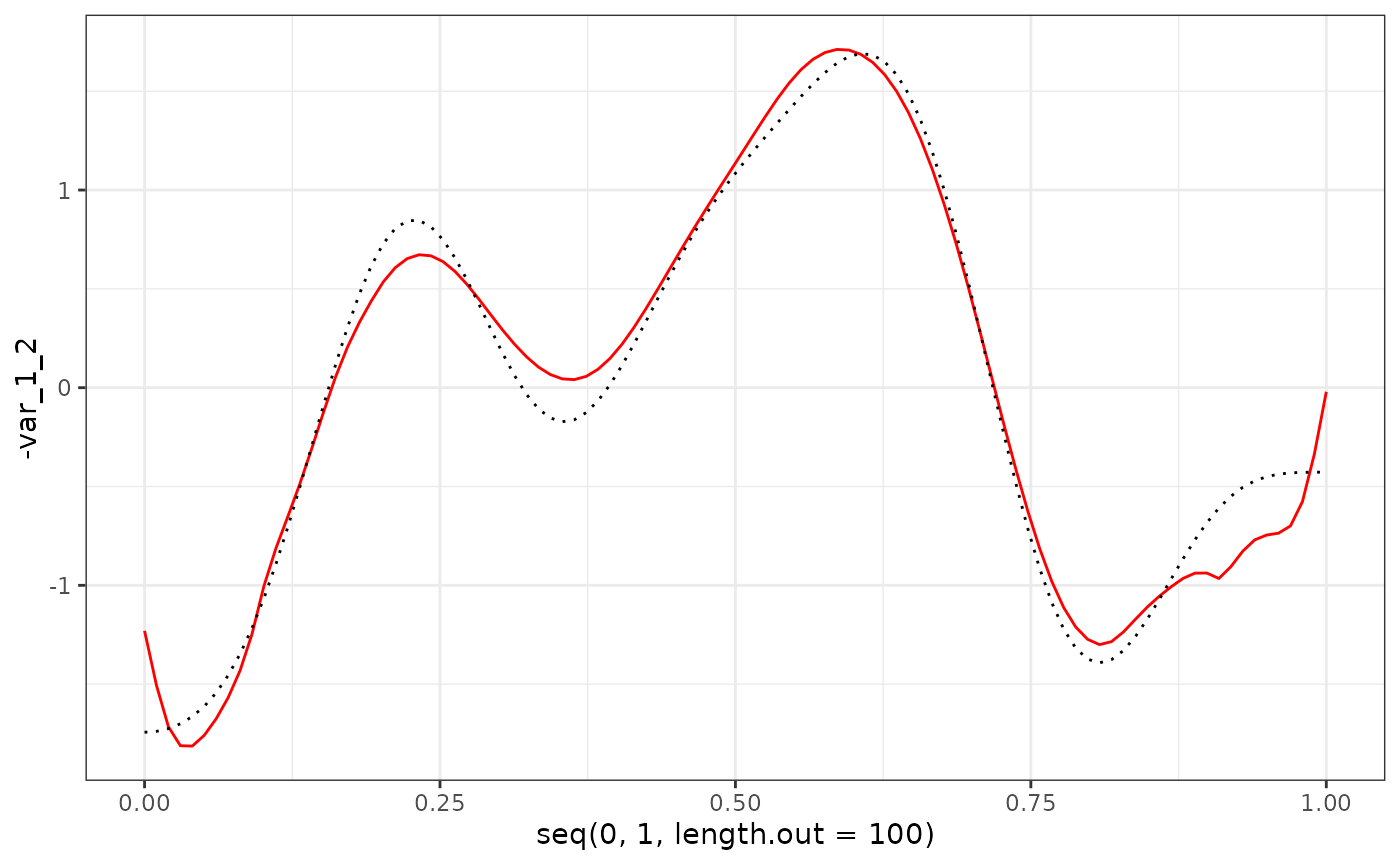
eigenf %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = seq(0, 1, length.out = 100), y = -var_1_3), col = "red") +
geom_line(aes(x = seq(0, 1, length.out = 100), y = phi_Y_df[1:100, 3]), col = "black", linetype = "dotted") +
theme_bw()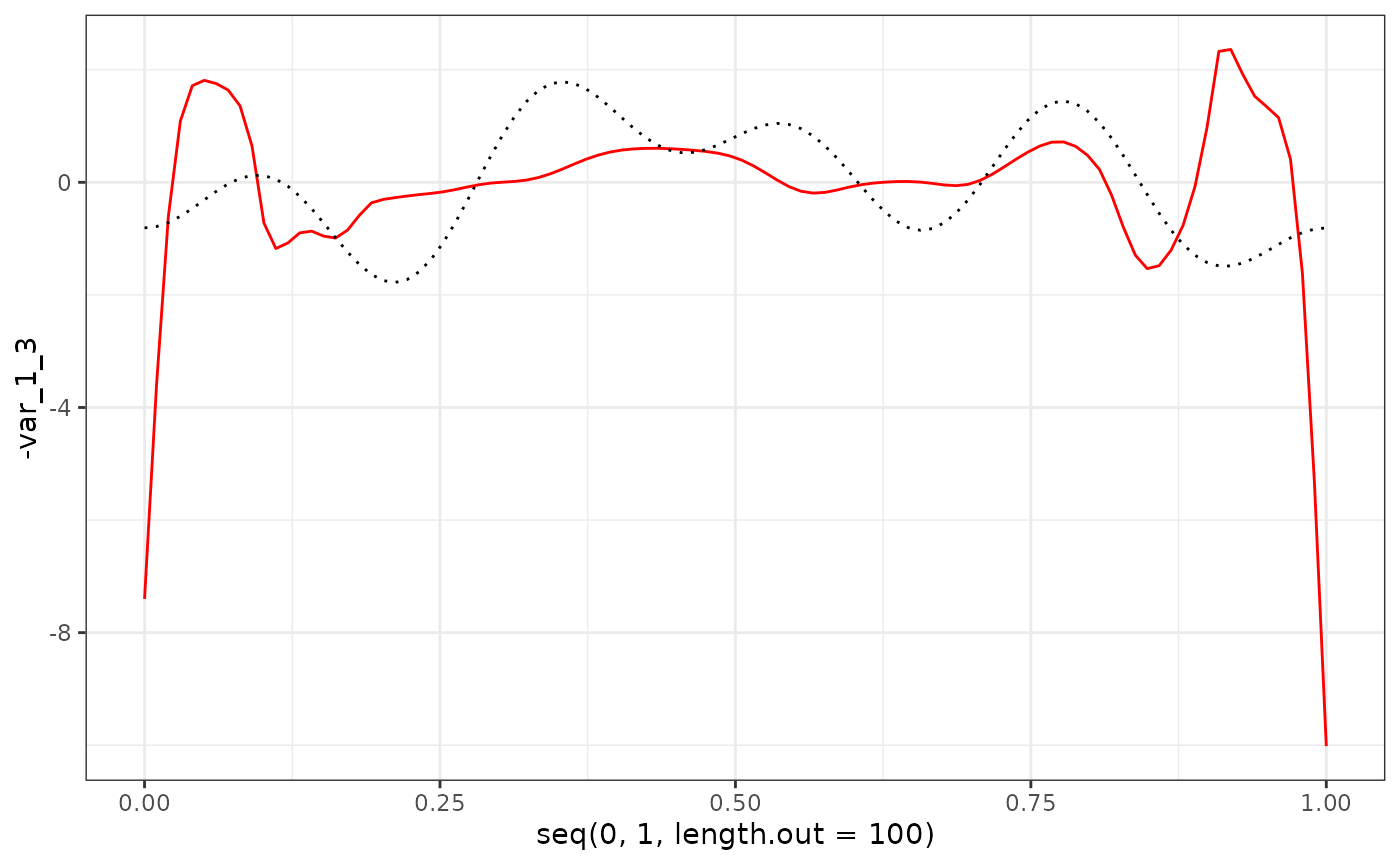
eigenf %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = seq(0, 1, length.out = 100), y = var_2_1), col = "red") +
geom_line(aes(x = seq(0, 1, length.out = 100), y = phi_Y_df[101:200, 1]), col = "black", linetype = "dotted") +
theme_bw()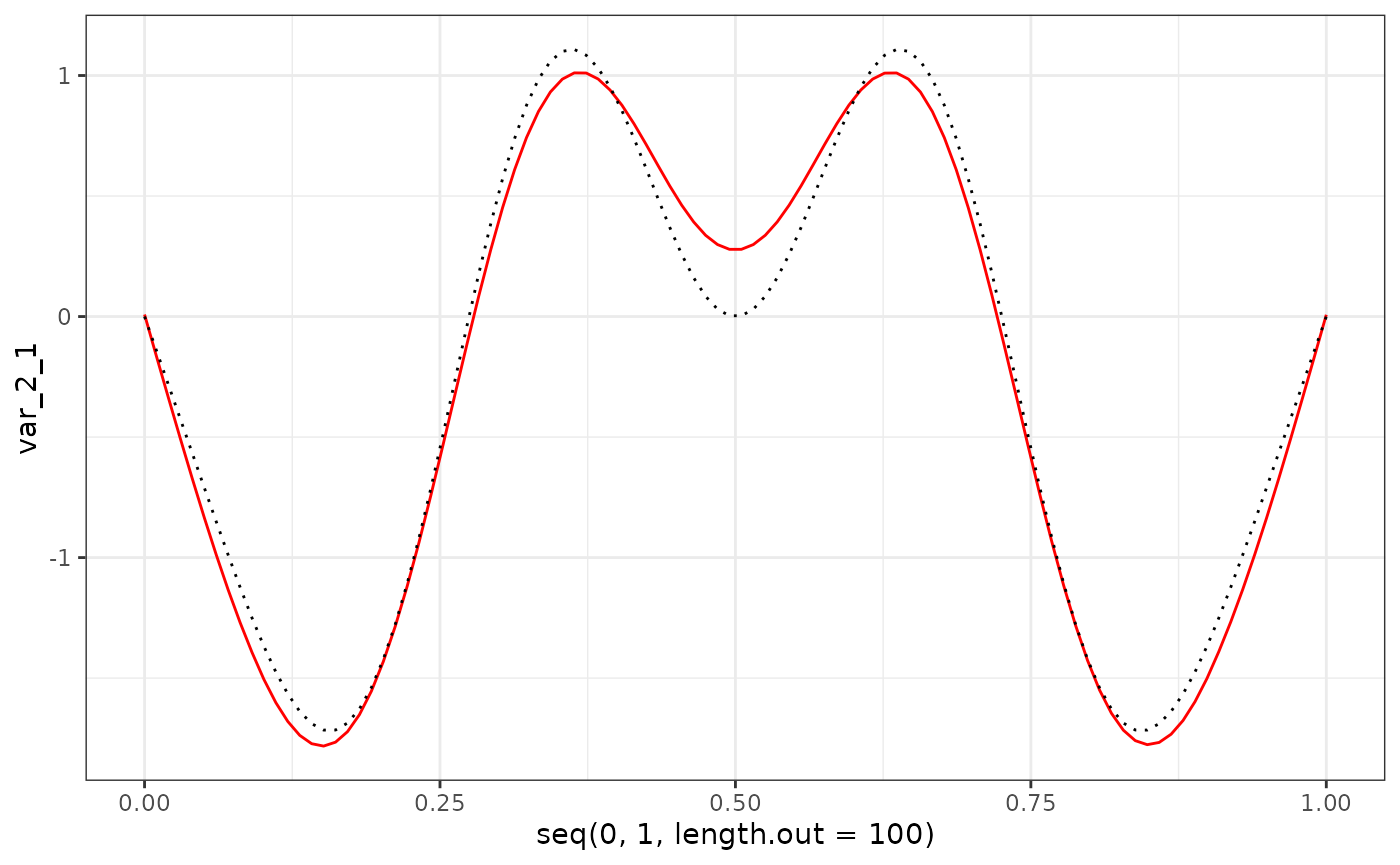
eigenf %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = seq(0, 1, length.out = 100), y = -var_2_2), col = "red") +
geom_line(aes(x = seq(0, 1, length.out = 100), y = phi_Y_df[101:200, 2]), col = "black", linetype = "dotted") +
theme_bw()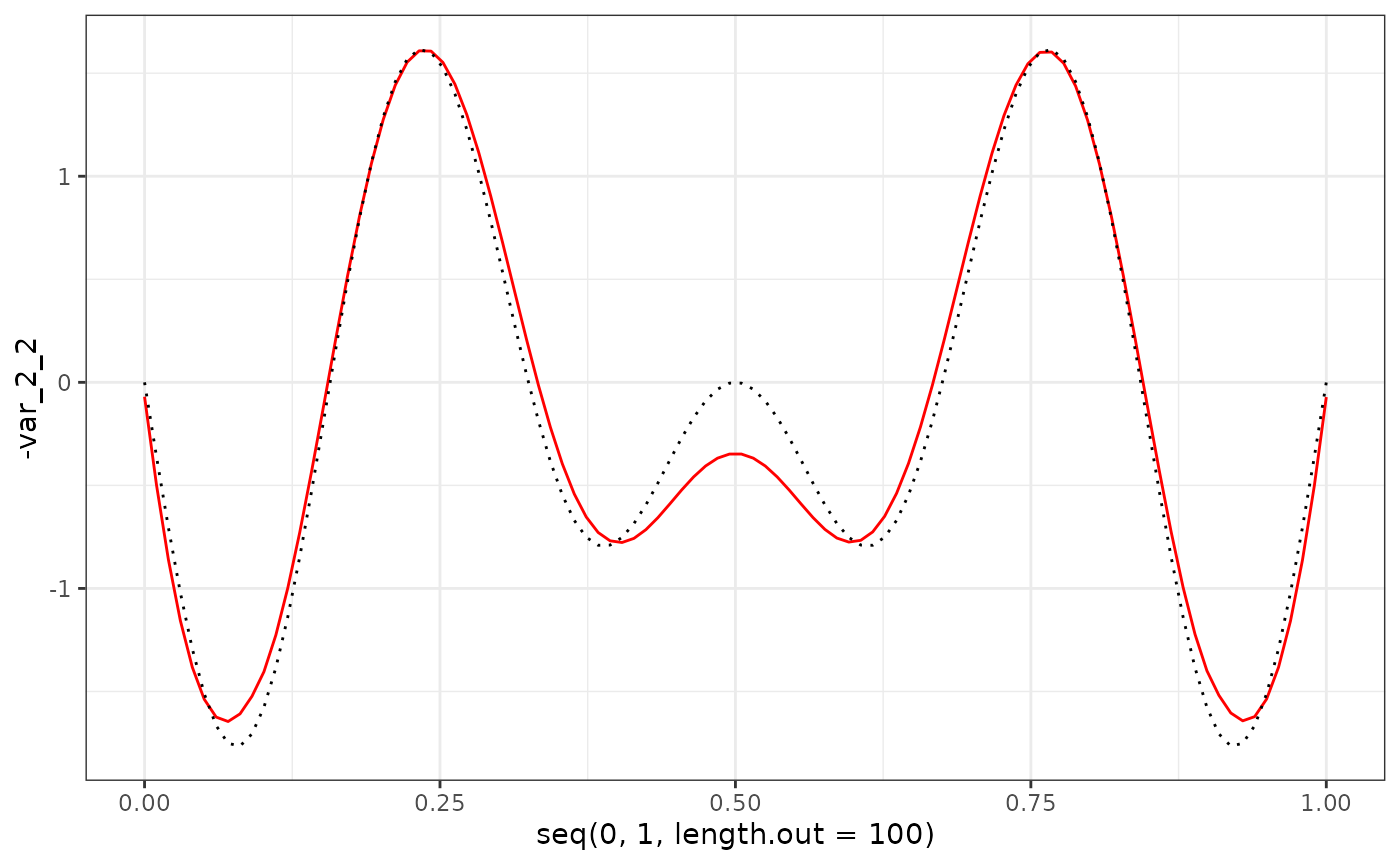
eigenf %>%
as.data.frame() %>%
ggplot() +
geom_line(aes(x = seq(0, 1, length.out = 100), y = -var_2_3), col = "red") +
geom_line(aes(x = seq(0, 1, length.out = 100), y = phi_Y_df[101:200, 3]), col = "black", linetype = "dotted") +
theme_bw()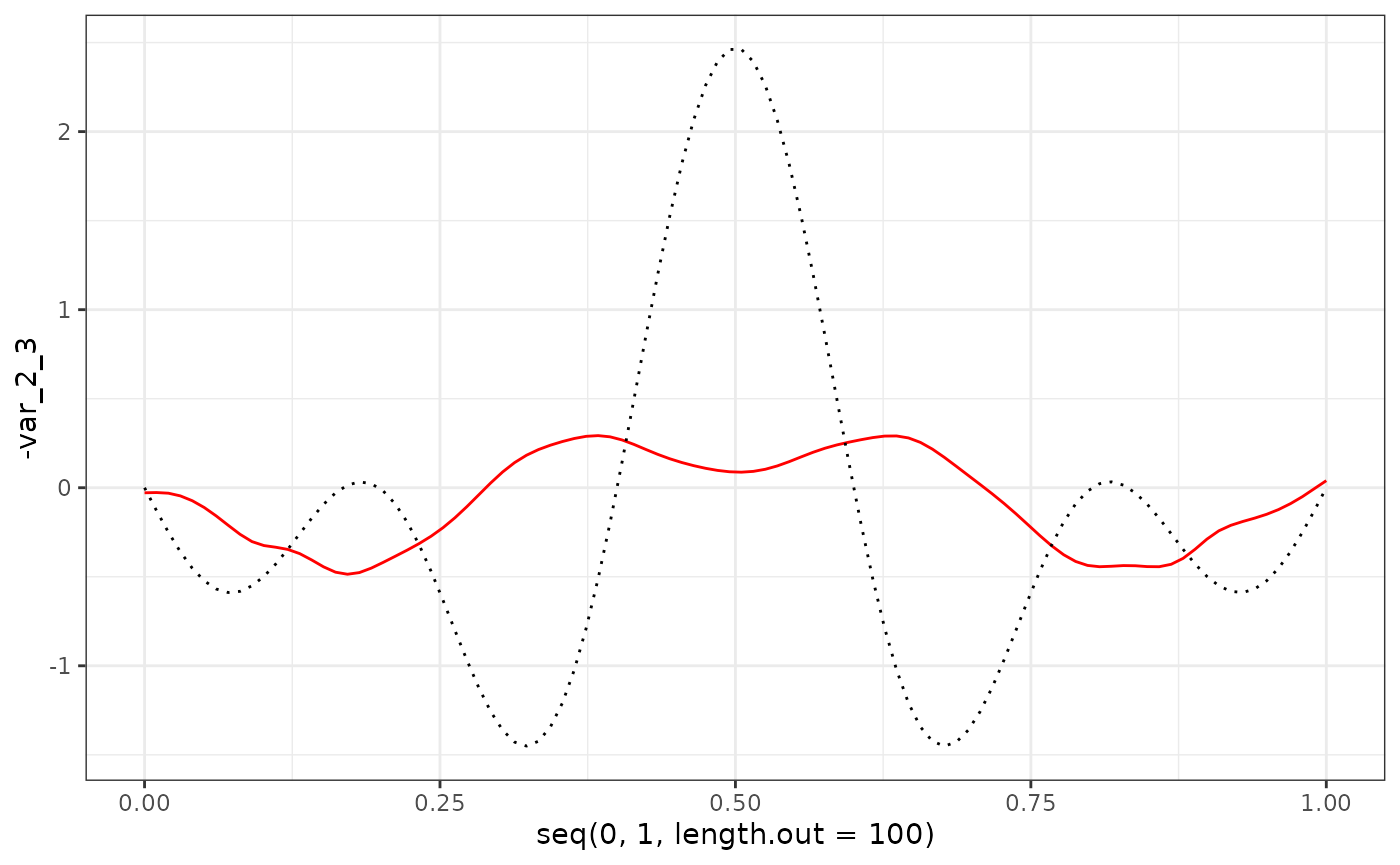
# eigens %>%
# ggplot(aes(x = est1, y = act1)) +
# geom_point() +
# geom_abline(intercept = 0, slope = 1, col = "red") +
# geom_smooth(method = "lm", se = F) +
# theme_bw()
#
# eigens %>%
# ggplot(aes(x = -est2, y = act2)) +
# geom_point() +
# geom_abline(intercept = 0, slope = 1, col = "red") +
# geom_smooth(method = "lm", se = F) +
# theme_bw()
#
# eigens %>%
# ggplot(aes(x = -est3, y = act3)) +
# geom_point() +
# geom_abline(intercept = 0, slope = 1, col = "red") +
# geom_smooth(method = "lm", se = F) +
# theme_bw()
eigens %>%
ggplot(aes(x = res_Y$xi[,1], y = popEig$pop_xiY[,1])) + # res_Y$xi[,1]
geom_point() +
geom_abline(intercept = 0, slope = 1, col = "red") +
geom_smooth(method = "lm", se = F) +
theme_bw()
#> `geom_smooth()` using formula = 'y ~ x'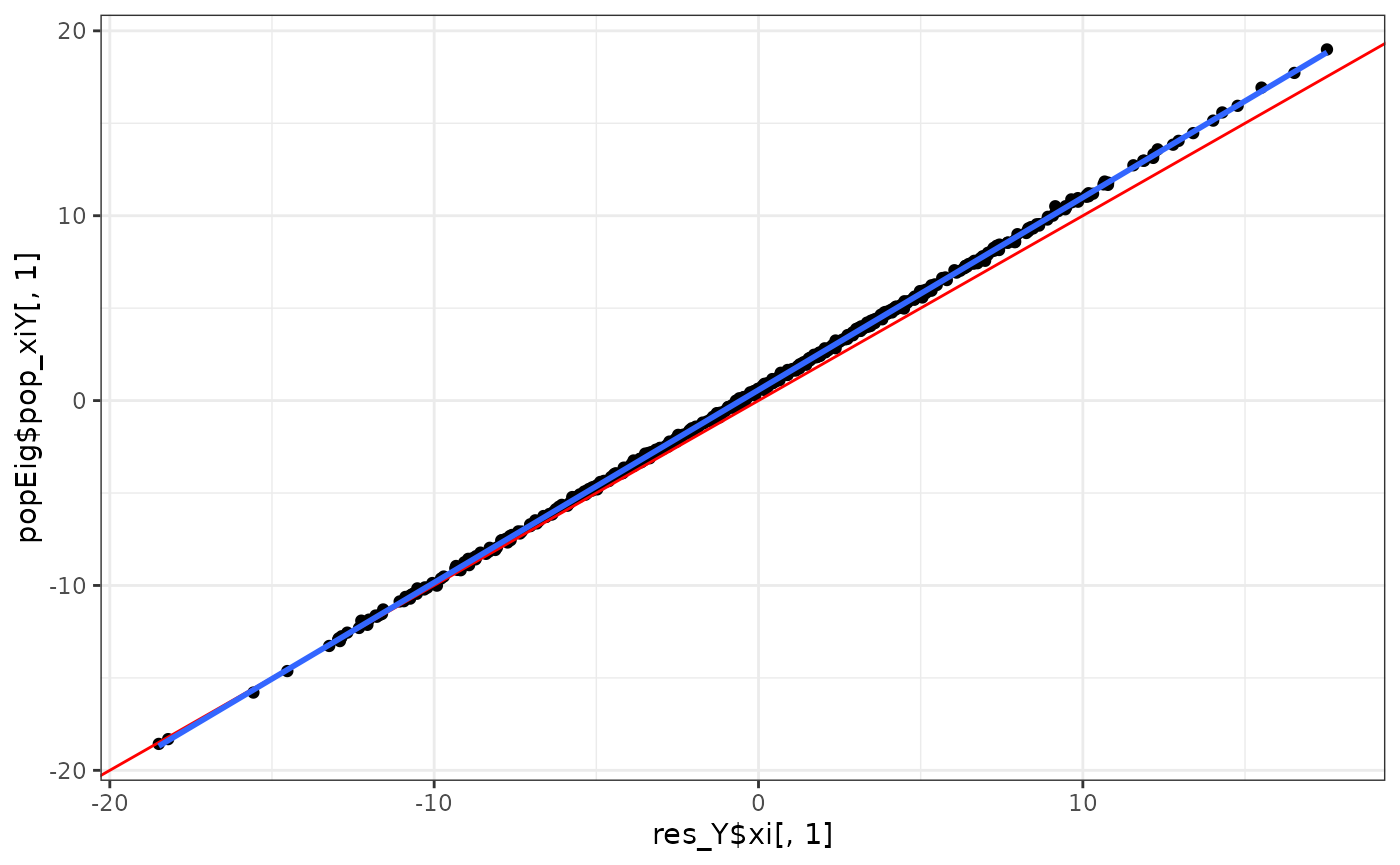
eigens %>%
ggplot(aes(x = -res_Y$xi[,2], y = popEig$pop_xiY[,2])) +
geom_point() +
geom_abline(intercept = 0, slope = 1, col = "red") +
geom_smooth(method = "lm", se = F) +
theme_bw()
#> `geom_smooth()` using formula = 'y ~ x'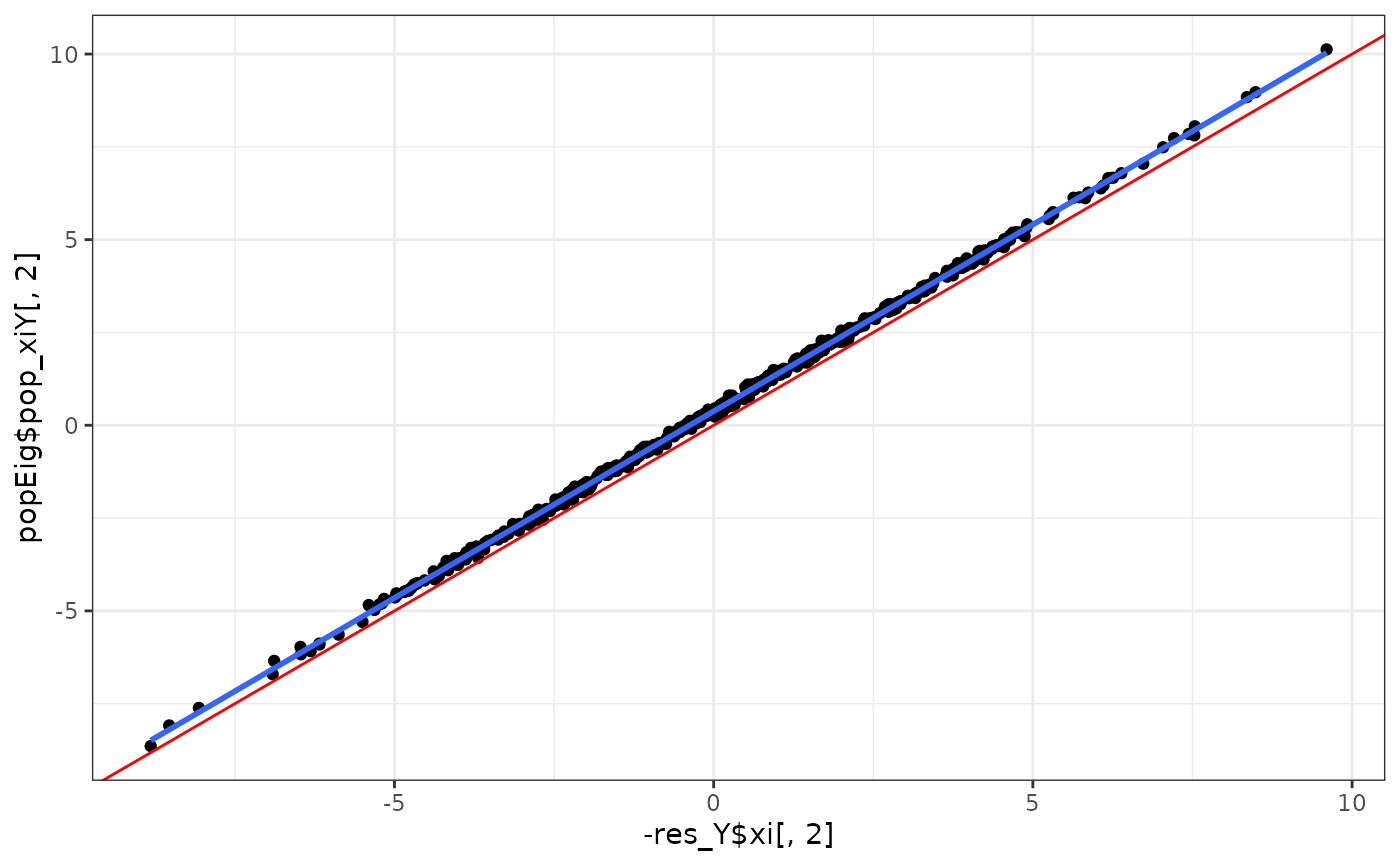
eigens %>%
ggplot(aes(x = res_Y$xi[,3], y = popEig$pop_xiY[,3])) +
geom_point() +
geom_abline(intercept = 0, slope = 1, col = "red") +
geom_smooth(method = "lm", se = F) +
theme_bw()
#> `geom_smooth()` using formula = 'y ~ x'
Regression
# response = res_Y$xi
# predictor = res$xi
# X_xi_pop <- Y$xi%*%Y$D
# mod <- lm(response ~ 0 + predictor)
Bhat = FunOnFun::funOnFun(res_Y, res)
# mod_sc_pop <- lm(popEig$pop_xiY ~ 0 + X_xi_pop)
Bestimand = FunOnFun::populationReg(X, popEig$pop_xiY)
Bestimand; Bhat$Bhat; popEig$pop_B
#> [,1] [,2] [,3]
#> xiX1 1.2084706 -0.22247723 0.0001157154
#> xiX2 -0.8022158 -0.93077996 -0.0001067740
#> xiX3 -0.1476355 -0.00865128 0.0287580183
#> [,1] [,2] [,3]
#> pred$xi1 1.1678595 0.21588536 -4.375704e-05
#> pred$xi2 0.7603738 -0.92345245 -6.280531e-05
#> pred$xi3 0.1516143 -0.02168497 6.384040e-03
#> [,1] [,2] [,3]
#> [1,] 1.2084720 -0.222477648 0.0001151556
#> [2,] -0.8022148 -0.930780571 -0.0001061576
#> [3,] -0.1476354 -0.008651756 0.02875671162D Comparison
# population = (eigen(popEig$pop_covY)$vectors[, 1:3]*sqrt(2*length(t))) %*% popEig$pop_B %*% t(X$phi)
population = FunOnFun::populationBeta(popEig$pop_covY, popEig$pop_B, t, X)
# estimated = res_Y$stacked_phi %*% mod$coefficients %*% t(res$stacked_phi)
estimated = FunOnFun::reconBeta(res_Y, res, Bhat$Bhat)
fig1 <- plotly::plot_ly(z = population,
type = "heatmap",
zmin = min(population),
zmax = max(population))
fig2 <- plotly::plot_ly(z = estimated,
type = "heatmap",
zmin = min(estimated),
zmax = max(estimated))
plotly::subplot(fig1, fig2)3D Comparison
estimated = estimated - 15
population_colorscale <- list(
list(0, "rgb(255, 0, 0)"),
list(1, "rgb(0, 255, 0)")
)
estimated_colorscale <- list(
list(0, "rgb(255, 0, 0)"),
list(1, "rgb(0, 255, 0)")
)
fig = plotly::plot_ly(showscale = F) %>%
plotly::add_surface(z = ~population,
cmin = min(population),
cmax = max(population),
colorscale = population_colorscale) %>%
plotly::add_surface(z = ~estimated,
cmin = min(estimated),
cmax = max(estimated),
colorscale = estimated_colorscale)
fig